| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-02-16 08:53:46 UTC |
|---|
| Update Date | 2023-02-21 17:15:49 UTC |
|---|
| HMDB ID | HMDB0001847 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Caffeine |
|---|
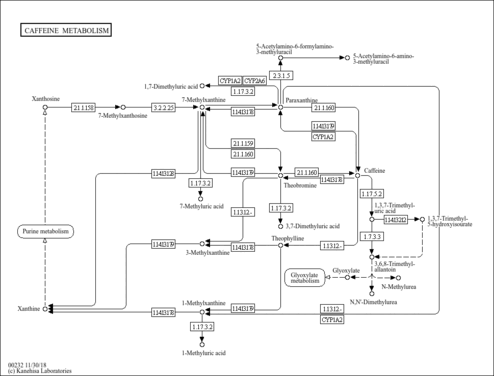
| Description | Caffeine is a methyl xanthine alkaloid that is also classified as a purine. Formally, caffeine belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. Caffeine is chemically related to the adenine and guanine bases of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). It is found in the seeds, nuts, or leaves of a number of plants native to Africa, East Asia and South America and helps to protect them against predator insects and to prevent germination of nearby seeds. The most well-known source of caffeine is the coffee bean. Caffeine is the most widely consumed psychostimulant drug in the world. 85% of American adults consumed some form of caffeine daily, consuming 164 mg on average. Caffeine is mostly is consumed in the form of coffee. Caffeine is a central nervous system stimulant that reduces fatigue and drowsiness. At normal doses, caffeine has variable effects on learning and memory, but it generally improves reaction time, wakefulness, concentration, and motor coordination. Caffeine is a proven ergogenic aid in humans. Caffeine improves athletic performance in aerobic (especially endurance sports) and anaerobic conditions. Moderate doses of caffeine (around 5 mg/kg) can improve sprint performance, cycling and running time trial performance, endurance and cycling power output (PMID: 32551869 ). At intake levels associated with coffee consumption, caffeine appears to exert most of its biological effects through the antagonism of the A1 and A2A subtypes of the adenosine receptor. Adenosine is an endogenous neuromodulator with mostly inhibitory effects, and adenosine antagonism by caffeine results in effects that are generally stimulatory. Some physiological effects associated with caffeine administration include central nervous system stimulation, acute elevation of blood pressure, increased metabolic rate, and diuresis. A number of in vitro and in vivo studies have demonstrated that caffeine modulates both innate and adaptive immune responses. For instance, studies indicate that caffeine and its major metabolite paraxanthine suppress neutrophil and monocyte chemotaxis, and also suppress production of the pro-inflammatory cytokine tumor necrosis factor (TNF) alpha from human blood. Caffeine has also been reported to suppress human lymphocyte function as indicated by reduced T-cell proliferation and impaired production of Th1 (interleukin [IL]-2 and interferon [IFN]-gamma), Th2 (IL-4, IL-5) and Th3 (IL-10) cytokines. Studies also indicate that caffeine suppresses antibody production. The evidence suggests that at least some of the immunomodulatory actions of caffeine are mediated via inhibition of cyclic adenosine monophosphate (cAMP)-phosphodiesterase (PDE), and consequential increase in intracellular cAMP concentrations. Overall, these studies indicate that caffeine, like other members of the methylxanthine family, is largely anti-inflammatory in nature, and based on the pharmacokinetics of caffeine, many of its immunomodulatory effects occur at concentrations that are relevant to normal human consumption. (PMID: 16540173 ). Caffeine is rapidly and almost completely absorbed in the stomach and small intestine and distributed to all tissues, including the brain. Caffeine metabolism occurs primarily in the liver, where the activity of the cytochrome P450 isoform CYP1A2 accounts for almost 95% of the primary metabolism of caffeine. CYP1A2-catalyzed 3-demethylation of caffeine results in the formation of 1,7-dimethylxanthine (paraxanthine). Paraxanthine may be demethylated by CYP1A2 to form 1-methylxanthine, which may be oxidized to 1-methyluric acid by xanthine oxidase. Paraxanthine may also be hydroxylated by CYP2A6 to form 1,7-dimethyluric acid, or acetylated by N-acetyltransferase 2 (NAT2) to form 5-acetylamino-6-formylamino-3-methyluracil, an unstable compound that may be deformylated nonenzymatically to form 5-acetylamino-6-amino-3-methyluracil. Caffeine concentrations in coffee beverages can be quite variable. A standard cup of coffee is often assumed to provide 100 mg of caffeine, but a recent analysis of 14 different specialty coffees purchased at coffee shops in the US found that the amount of caffeine in 8 oz (=240 ml) of brewed coffee ranged from 72 to 130 mg. Caffeine in espresso coffees ranged from 58 to 76 mg in a single shot. (PMID: 16507475 ). |
|---|
| Structure | CN1C=NC2=C1C(=O)N(C)C(=O)N2C InChI=1S/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)8(14)11(6)2/h4H,1-3H3 |
|---|
| Synonyms | | Value | Source |
|---|
| 1,3,7-Trimethyl-2,6-dioxopurine | ChEBI | | 1,3,7-Trimethylpurine-2,6-dione | ChEBI | | 1,3,7-Trimethylxanthine | ChEBI | | 1-Methyltheobromine | ChEBI | | 3,7-Dihydro-1,3,7-trimethyl-1H-purin-2,6-dion | ChEBI | | 7-Methyltheophylline | ChEBI | | Anhydrous caffeine | ChEBI | | Cafeina | ChEBI | | Cafeine | ChEBI | | Coffein | ChEBI | | Guaranine | ChEBI | | Koffein | ChEBI | | Mateina | ChEBI | | Methyltheobromine | ChEBI | | Teina | ChEBI | | Thein | ChEBI | | Theine | ChEBI | | Respia | Kegg | | 1,3,7-Trimethyl-3,7-dihydro-1H-purine-2,6-dione | HMDB | | 1-Methyl-theobromine | HMDB | | 3,7-Dihydro-1,3,7-trimethyl-1H-purine-2,6-dione | HMDB | | 7-Methyl theophylline | HMDB | | Anhydrous caffeine (JP15) | HMDB | | Hycomine | HMDB | | Lanorinal | HMDB | | Methyltheobromide | HMDB | | Methylxanthine theophylline | HMDB | | Monohydrate caffeine | HMDB | | Propoxyphene | HMDB | | Merck dura brand OF caffeine | HMDB | | Thompson brand 1 OF caffeine | HMDB | | Bristol-myers squibb brand OF caffeine | HMDB | | Caffedrine | HMDB | | Dexitac | HMDB | | Percoffedrinol N | HMDB | | Pierre fabre brand OF caffeine | HMDB | | Republic drug brand OF caffeine | HMDB | | Thompson brand 2 OF caffeine | HMDB | | Vivarin | HMDB | | Coffeinum N | HMDB | | Coffeinum purrum | HMDB | | Durvitan | HMDB | | GlaxoSmithKline brand OF caffeine | HMDB | | No doz | HMDB | | Percutaféine | HMDB | | Quick-pep | HMDB | | Seid brand OF caffeine | HMDB | | Berlin-chemie brand OF caffeine | HMDB | | Passauer brand OF caffeine | HMDB | | Quick pep | HMDB | | QuickPep | HMDB |
|
|---|
| Chemical Formula | C8H10N4O2 |
|---|
| Average Molecular Weight | 194.1906 |
|---|
| Monoisotopic Molecular Weight | 194.080375584 |
|---|
| IUPAC Name | 1,3,7-trimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione |
|---|
| Traditional Name | caffeine |
|---|
| CAS Registry Number | 58-08-2 |
|---|
| SMILES | CN1C=NC2=C1C(=O)N(C)C(=O)N2C |
|---|
| InChI Identifier | InChI=1S/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)8(14)11(6)2/h4H,1-3H3 |
|---|
| InChI Key | RYYVLZVUVIJVGH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- Purinone
- 6-oxopurine
- Alkaloid or derivatives
- Pyrimidone
- Pyrimidine
- N-substituted imidazole
- Heteroaromatic compound
- Vinylogous amide
- Imidazole
- Azole
- Urea
- Lactam
- Azacycle
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | Biological locationRoute of exposureSourceEndogenousExogenousFood- Food (HMDB: HMDB0001847)
Animal originVegetableHerb and spiceNutFruitCereal and cereal productPulseGourdSoyTeaBeverageAquatic originConfectioneryMilk and milk productOther milk productFermented milkFermented milk productUnfermented milk- Milk (Other mammals) (FooDB: FOOD00690)
- Milk (Human) (FooDB: FOOD00666)
- Milk (Cow) (FooDB: FOOD00618)
- Cow milk, pasteurized, vitamin A + D added, 0% fat (FooDB: FOOD00889)
- Cow milk, pasteurized, vitamin A + D added, 1% fat (FooDB: FOOD00890)
- Cow milk, pasteurized, vitamin A + D added, 2% fat (FooDB: FOOD00891)
- Cow milk, pasteurized, vitamin D added, 3.25% fat (FooDB: FOOD00892)
Fat and oilCocoa and cocoa productBaking goodBaby foodUnclassified food or beverageDishCoffee and coffee productSnackEgg
SyntheticEnvironmental |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 238 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 21.6 mg/mL at 25 °C | Not Available | | LogP | -0.07 | HANSCH,C ET AL. (1995) |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (0 TMS) | splash10-0536-3900000000-a9e112713ffae6dabdaa | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (0 TMS) | splash10-0536-2900000000-8cdcd005b2e7622a02a3 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-052f-0900000000-f1084acfddb240696073 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-05nf-6900000000-8670a644cee5d9de78d4 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-MS (Non-derivatized) | splash10-0536-3900000000-4430852b279a72e34822 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine EI-B (Non-derivatized) | splash10-0006-0900000000-51898e93480e848d7da1 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine CI-B (Non-derivatized) | splash10-0002-0900000000-2aed5d425b6a95add5db | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine EI-B (Non-derivatized) | splash10-0a4l-4900000000-3ff72dace6687d242f1f | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine EI-B (Non-derivatized) | splash10-0006-1900000000-2ba1fae6e27c7b836984 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine CI-B (Non-derivatized) | splash10-0002-0900000000-fd859aeb416e320d6379 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Non-derivatized) | splash10-0536-3900000000-a9e112713ffae6dabdaa | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Non-derivatized) | splash10-0536-2900000000-8cdcd005b2e7622a02a3 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Non-derivatized) | splash10-052f-0900000000-f1084acfddb240696073 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-EI-TOF (Non-derivatized) | splash10-05nf-6900000000-8670a644cee5d9de78d4 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Caffeine GC-MS (Non-derivatized) | splash10-0536-3900000000-4430852b279a72e34822 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Caffeine GC-MS (Non-derivatized) - 70eV, Positive | splash10-052r-0900000000-41f36d541d34d2088964 | 2017-07-27 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Caffeine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-052f-8900000000-68b5e9aa3404fb3d8d3a | 2014-09-20 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-0006-0900000000-447fc72b2c709e2e18a9 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-000i-1900000000-5e3b29de16ad91c79fe0 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-0006-9100000000-d6f6c52ac36c8f25a500 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine EI-B (HITACHI M-80) , Positive-QTOF | splash10-0006-0900000000-cddd24399d942b1ac97c | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine CI-B (Unknown) , Positive-QTOF | splash10-0002-0900000000-2aed5d425b6a95add5db | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine EI-B (HITACHI M-60) , Positive-QTOF | splash10-0a4l-4900000000-3ff72dace6687d242f1f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine EI-B (HITACHI M-68) , Positive-QTOF | splash10-0006-1900000000-2ba1fae6e27c7b836984 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine CI-B (HITACHI M-60) , Positive-QTOF | splash10-0002-0900000000-63b9ef42a3e8d59e9997 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive-QTOF | splash10-0002-0900000000-185b3d97d8857a0f269d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive-QTOF | splash10-0002-0900000000-f8a0c0dd9f5c4a272eaf | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive-QTOF | splash10-000i-1900000000-dd8e35226d0704aa657d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive-QTOF | splash10-01x9-9800000000-70e3b0eb52481c39d191 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive-QTOF | splash10-001l-9100000000-6d428a5571beb0e3fed4 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive-QTOF | splash10-0002-0900000000-98bec16f898808c3de68 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive-QTOF | splash10-0002-0900000000-b112e4e059e1ecf98c5f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine DI-ESI-qTof , Positive-QTOF | splash10-00dr-0900000000-42c6f8fc7b924e9c64f3 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-qTof , Positive-QTOF | splash10-000i-4900000000-a60a480f1340558740a2 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-000i-0900000000-695d910d49fc0beb1d54 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-0002-0900000000-094879886a2e72bf0c56 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-0002-0900000000-fa38c865089a3a05f287 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-000b-0900000000-0e82732a924c974dd0c8 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-000i-0900000000-bfc94c8091471847482b | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-000i-1900000000-c8fcf16986c494898203 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-000i-3900000000-9569e0552abb7ebd145a | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Caffeine LC-ESI-ITFT , positive-QTOF | splash10-0002-0900000000-3a924abd44877050c1c9 | 2017-09-14 | HMDB team, MONA | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, CDCl3, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Disease References | | Head injury |
|---|
- Sachse KT, Jackson EK, Wisniewski SR, Gillespie DG, Puccio AM, Clark RS, Dixon CE, Kochanek PM: Increases in cerebrospinal fluid caffeine concentration are associated with favorable outcome after severe traumatic brain injury in humans. J Cereb Blood Flow Metab. 2008 Feb;28(2):395-401. Epub 2007 Aug 8. [PubMed:17684518 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Metastatic melanoma |
|---|
- Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY: Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017 Oct;19(10):848-855. doi: 10.1016/j.neo.2017.08.004. Epub 2017 Sep 15. [PubMed:28923537 ]
| | Asthma |
|---|
- Zydron M, Baranowski J, Baranowska I: Separation, pre-concentration, and HPLC analysis of methylxanthines in urine samples. J Sep Sci. 2004 Oct;27(14):1166-72. [PubMed:15537072 ]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
|
|
|---|
| General References | - Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Miyake Y, Sakaguchi K, Iwasaki Y, Ikeda H, Makino Y, Kobashi H, Araki Y, Ando M, Kita K, Shiratori Y: New prognostic scoring model for liver transplantation in patients with non-acetaminophen-related fulminant hepatic failure. Transplantation. 2005 Oct 15;80(7):930-6. [PubMed:16249741 ]
- Wilkinson SC, Maas WJ, Nielsen JB, Greaves LC, van de Sandt JJ, Williams FM: Interactions of skin thickness and physicochemical properties of test compounds in percutaneous penetration studies. Int Arch Occup Environ Health. 2006 May;79(5):405-13. Epub 2006 Jan 25. [PubMed:16435152 ]
- Spiller HA, Winter ML, Klein-Schwartz W, Bangh SA: Efficacy of activated charcoal administered more than four hours after acetaminophen overdose. J Emerg Med. 2006 Jan;30(1):1-5. [PubMed:16434328 ]
- Ayotte P, Dewailly E, Lambert GH, Perkins SL, Poon R, Feeley M, Larochelle C, Pereg D: Biomarker measurements in a coastal fish-eating population environmentally exposed to organochlorines. Environ Health Perspect. 2005 Oct;113(10):1318-24. [PubMed:16203240 ]
- Shah S, Budev M, Blazey H, Fairbanks K, Mehta A: Hepatic veno-occlusive disease due to tacrolimus in a single-lung transplant patient. Eur Respir J. 2006 May;27(5):1066-8. [PubMed:16707401 ]
- Larson AM, Polson J, Fontana RJ, Davern TJ, Lalani E, Hynan LS, Reisch JS, Schiodt FV, Ostapowicz G, Shakil AO, Lee WM: Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005 Dec;42(6):1364-72. [PubMed:16317692 ]
- Septer S, Thompson ES, Willemsen-Dunlap A: Anesthesia concerns for children with tuberous sclerosis. AANA J. 2006 Jun;74(3):219-25. [PubMed:16786916 ]
- Horrigan LA, Kelly JP, Connor TJ: Immunomodulatory effects of caffeine: friend or foe? Pharmacol Ther. 2006 Sep;111(3):877-92. Epub 2006 Mar 15. [PubMed:16540173 ]
- Rodrigues IM, Klein LC: Boiled or filtered coffee? Effects of coffee and caffeine on cholesterol, fibrinogen and C-reactive protein. Toxicol Rev. 2006;25(1):55-69. [PubMed:16856769 ]
- Lamarine RJ: Selected health and behavioral effects related to the use of caffeine. J Community Health. 1994 Dec;19(6):449-66. [PubMed:7844249 ]
- James JE: Critical review of dietary caffeine and blood pressure: a relationship that should be taken more seriously. Psychosom Med. 2004 Jan-Feb;66(1):63-71. [PubMed:14747639 ]
- Higdon JV, Frei B: Coffee and health: a review of recent human research. Crit Rev Food Sci Nutr. 2006;46(2):101-23. [PubMed:16507475 ]
- Nehlig A, Daval JL, Debry G: Caffeine and the central nervous system: mechanisms of action, biochemical, metabolic and psychostimulant effects. Brain Res Brain Res Rev. 1992 May-Aug;17(2):139-70. [PubMed:1356551 ]
- Benjamin LT Jr, Rogers AM, Rosenbaum A: Coca-Cola, caffeine, and mental deficiency: Harry Hollingworth and the Chattanooga trial of 1911. J Hist Behav Sci. 1991 Jan;27(1):42-55. [PubMed:2010614 ]
- Nathanson JA: Caffeine and related methylxanthines: possible naturally occurring pesticides. Science. 1984 Oct 12;226(4671):184-7. [PubMed:6207592 ]
- Smit HJ, Gaffan EA, Rogers PJ: Methylxanthines are the psycho-pharmacologically active constituents of chocolate. Psychopharmacology (Berl). 2004 Nov;176(3-4):412-9. Epub 2004 May 5. [PubMed:15549276 ]
- Haskell CF, Kennedy DO, Wesnes KA, Milne AL, Scholey AB: A double-blind, placebo-controlled, multi-dose evaluation of the acute behavioural effects of guarana in humans. J Psychopharmacol. 2007 Jan;21(1):65-70. Epub 2006 Mar 13. [PubMed:16533867 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
- Ferreira TT, da Silva JVF, Bueno NB: Effects of caffeine supplementation on muscle endurance, maximum strength, and perceived exertion in adults submitted to strength training: a systematic review and meta-analyses. Crit Rev Food Sci Nutr. 2021;61(15):2587-2600. doi: 10.1080/10408398.2020.1781051. Epub 2020 Jun 18. [PubMed:32551869 ]
|
|---|