| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:08 UTC |
|---|
| HMDB ID | HMDB0001070 |
|---|
| Secondary Accession Numbers | - HMDB0006257
- HMDB01070
- HMDB06257
|
|---|
| Metabolite Identification |
|---|
| Common Name | Octanoyl-CoA |
|---|
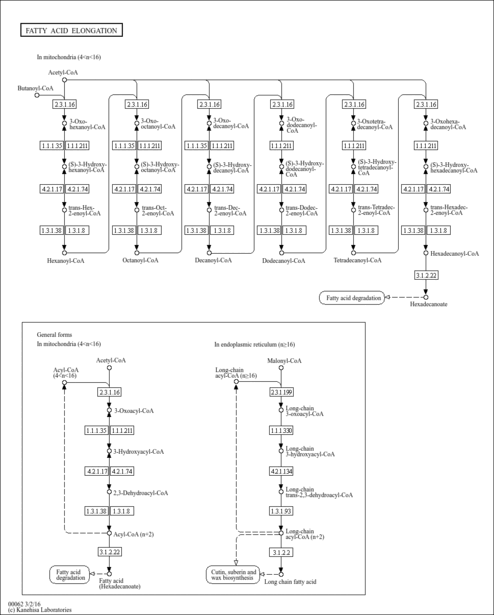
| Description | Octanoyl-CoA is a substrate for Trifunctional enzyme beta subunit (mitochondrial), Acyl-coenzyme A oxidase 1 (peroxisomal), 3-ketoacyl-CoA thiolase (mitochondrial), 3-ketoacyl-CoA thiolase (peroxisomal), Nuclear receptor-binding factor 1, Acyl-CoA dehydrogenase (long-chain specific, mitochondrial), Acyl-coenzyme A oxidase 3 (peroxisomal), HPDHase, Acyl-CoA dehydrogenase (medium-chain specific, mitochondrial), Acyl-coenzyme A oxidase 2 (peroxisomal) and Peroxisomal carnitine O-octanoyltransferase. |
|---|
| Structure | CCCCCCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OCC1OC(C(O)C1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 InChI=1S/C29H50N7O17P3S/c1-4-5-6-7-8-9-20(38)57-13-12-31-19(37)10-11-32-27(41)24(40)29(2,3)15-50-56(47,48)53-55(45,46)49-14-18-23(52-54(42,43)44)22(39)28(51-18)36-17-35-21-25(30)33-16-34-26(21)36/h16-18,22-24,28,39-40H,4-15H2,1-3H3,(H,31,37)(H,32,41)(H,45,46)(H,47,48)(H2,30,33,34)(H2,42,43,44) |
|---|
| Synonyms | | Value | Source |
|---|
| Capryloyl-CoA | HMDB | | Capryloyl-coenzyme A | HMDB | | Octanoyl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C29H50N7O17P3S |
|---|
| Average Molecular Weight | 893.73 |
|---|
| Monoisotopic Molecular Weight | 893.219673435 |
|---|
| IUPAC Name | {[5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({[hydroxy({3-hydroxy-2,2-dimethyl-3-[(2-{[2-(octanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]propoxy})phosphoryl]oxy})phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | octanoyl-coenzyme A |
|---|
| CAS Registry Number | 1264-52-4 |
|---|
| SMILES | CCCCCCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OCC1OC(C(O)C1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 |
|---|
| InChI Identifier | InChI=1S/C29H50N7O17P3S/c1-4-5-6-7-8-9-20(38)57-13-12-31-19(37)10-11-32-27(41)24(40)29(2,3)15-50-56(47,48)53-55(45,46)49-14-18-23(52-54(42,43)44)22(39)28(51-18)36-17-35-21-25(30)33-16-34-26(21)36/h16-18,22-24,28,39-40H,4-15H2,1-3H3,(H,31,37)(H,32,41)(H,45,46)(H,47,48)(H2,30,33,34)(H2,42,43,44) |
|---|
| InChI Key | KQMZYOXOBSXMII-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 2,3,4-saturated fatty acyl coas. These are acyl-CoAs carrying a 2,3,4-saturated fatty acyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | 2,3,4-saturated fatty acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 4.55 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 11.1635 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 9.22 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 453.4 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 1804.0 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 147.1 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 130.3 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 160.7 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 111.5 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 409.4 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 517.7 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 954.3 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 749.1 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 461.7 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 746.1 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 274.0 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 248.9 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 395.1 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 215.2 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 270.5 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | 2022-08-06 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 10V, Positive-QTOF | splash10-000i-1912000220-9b11519b56edd2ee2e36 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 20V, Positive-QTOF | splash10-000i-0933000000-e284a1608276fcd6d82a | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 40V, Positive-QTOF | splash10-000i-1920000000-55d461927db02990f66b | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 10V, Negative-QTOF | splash10-0059-3911030340-f67f4b81132c4f964314 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 20V, Negative-QTOF | splash10-003r-2911010010-808f2ca30f372f07c257 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 40V, Negative-QTOF | splash10-057i-6900100000-fa25044ba9d73d317f9e | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 10V, Positive-QTOF | splash10-0006-0000000090-f44680cdef99cbcf2e1a | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 20V, Positive-QTOF | splash10-004l-0010100890-e02b1335b66645baa779 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 40V, Positive-QTOF | splash10-000i-0119000000-ddc6371111371aea35ac | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 10V, Negative-QTOF | splash10-0006-0000000090-b13a15ce3a6a0b95fb75 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 20V, Negative-QTOF | splash10-0096-5100302590-b8c368142a42ee60f3a4 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Octanoyl-CoA 40V, Negative-QTOF | splash10-00p3-6105712940-045236c3e4d4e3a8e5d1 | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum |
|
|---|
| General References | - Partanen ST, Novikov DK, Popov AN, Mursula AM, Hiltunen JK, Wierenga RK: The 1.3 A crystal structure of human mitochondrial Delta3-Delta2-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group. J Mol Biol. 2004 Sep 24;342(4):1197-208. [PubMed:15351645 ]
- Schowalter DB, Matern D, Vockley J: In vitro correction of medium chain acyl CoA dehydrogenase deficiency with a recombinant adenoviral vector. Mol Genet Metab. 2005 Jun;85(2):88-95. Epub 2005 Mar 19. [PubMed:15896652 ]
- Gargus JJ, Boyle K, Bocian M, Roe DS, Vianey-Saban C, Roe CR: Respiratory complex II defect in siblings associated with a symptomatic secondary block in fatty acid oxidation. J Inherit Metab Dis. 2003;26(7):659-70. [PubMed:14707514 ]
- Parker AR: Binding of the human "electron transferring flavoprotein" (ETF) to the medium chain acyl-CoA dehydrogenase (MCAD) involves an arginine and histidine residue. J Enzyme Inhib Med Chem. 2003 Oct;18(5):453-62. [PubMed:14692513 ]
|
|---|