Showing metabocard for Adenosylcobalamin (HMDB0002086)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 5.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Expected but not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2006-05-22 14:17:35 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2022-03-07 02:49:12 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0002086 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Adenosylcobalamin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
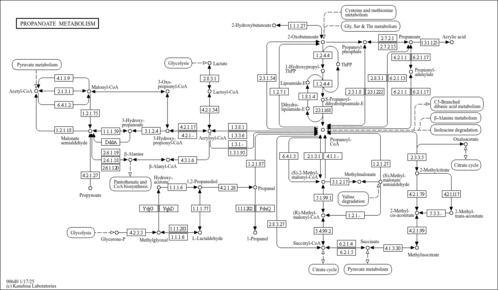
| Description | Adenosylcobalamin is one of two metabolically active forms synthesized upon ingestion of vitamin B12 and is the predominant form in the liver; it acts as a coenzyme in the reaction catalyzed by methylmalonyl-CoA mutase. A cobalamin (cbl) derivative in which the substituent is deoxyadenosyl. It is one of two metabolically active forms synthesized upon ingestion of vitamin B12 and is the predominant form in the liver; it acts as a coenzyme in the reaction catalyzed by methylmalonyl-CoA mutase (MCM; E.C. 5.4.99.2). Inborn errors of vitamin B12 metabolism are autosomal recessive disorders and have been classified into nine distinct complementation classes. Disorders affecting adenosylcobalamin cause methylmalonic acidemia and metabolic acidosis. Methylmalonyl-CoA mutase catalyzes the conversion of L-methylmalonyl-CoA to succinyl-CoA and uses adenosylcobalamin (AdoCbl) as a cofactor. Cbl must be transported into mitochondria, reduced and adenosylated before it can be utilized by MCM. (PMID: 17011224 ). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | MOL for HMDB0002086 (Adenosylcobalamin)
Mrv1652305171818062D
110124 0 0 1 0 999 V2000
0.2847 3.6890 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.1654 2.9976 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.5048 2.5166 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.0023 3.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4991 3.9582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1479 4.7047 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.3212 3.8891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2545 1.7305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.4032 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7433 1.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5281 0.9263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8944 1.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4840 2.9616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6968 1.8883 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.0455 2.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8655 2.8503 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.7212 2.3422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1966 3.6059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0166 3.6970 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
4.3840 4.3306 0.0000 O 0 5 0 0 0 0 0 0 0 0 0 0
4.7392 4.5486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3477 4.4526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1677 4.5436 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
6.5760 5.2605 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.3030 6.0918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5106 6.9125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.3839 5.0937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.4750 4.2737 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.7233 3.9338 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
6.4005 3.1540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.3072 3.8479 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
8.9436 4.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3838 3.6131 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
9.0909 2.8787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6393 3.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3798 4.2781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2019 4.6166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5718 4.4449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5697 5.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0234 3.8286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.2829 3.0455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2580 2.9728 0.0000 Co 0 7 0 0 0 0 0 0 0 0 0 0
0.4677 2.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2815 1.9756 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6884 1.2579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4967 1.4231 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.5893 2.2429 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.5360 2.3059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8383 2.5844 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.1530 3.4299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1054 0.8663 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.9137 1.0315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3206 0.3138 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.7637 -0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8435 -1.1161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5945 -1.4576 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.1723 -1.5958 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.4213 -1.2543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3415 -0.4332 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.0127 0.0465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8300 2.5088 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.5705 1.7257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3198 0.9709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3332 0.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1257 0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5424 1.5067 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
-3.3483 1.3305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0206 2.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1117 2.9745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3600 3.3145 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
-3.4510 4.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5612 4.7169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6536 5.5395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7362 4.7217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4767 3.9386 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
-0.6517 3.9434 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.0525 4.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4013 4.7295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.3381 5.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2795 5.4046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0067 6.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8926 6.0577 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
1.2271 6.8246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0716 5.2105 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.1957 6.0863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5085 6.9296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5568 7.8132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1436 8.3984 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.7983 8.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2590 4.3012 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-4.0664 5.1588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9595 4.9013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4260 5.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3187 6.5417 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-6.2630 5.8135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6673 3.5843 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-5.4893 3.3133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1274 2.7045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.0020 2.6843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.4826 1.9987 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-7.5621 3.3516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4298 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9067 -0.2118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2789 0.6327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6741 0.1784 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.6523 -0.6852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8365 -1.5532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4025 -2.3163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5713 -2.4256 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-2.8229 -3.0752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2 1 1 6 0 0 0
2 3 1 0 0 0 0
3 4 1 6 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
5 7 2 0 0 0 0
3 8 1 0 0 0 0
8 9 1 6 0 0 0
8 10 1 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
12 14 1 0 0 0 0
14 15 1 0 0 0 0
16 15 1 0 0 0 0
16 17 1 6 0 0 0
16 18 1 0 0 0 0
18 19 1 0 0 0 0
19 20 1 0 0 0 0
19 21 2 0 0 0 0
19 22 1 0 0 0 0
23 22 1 6 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 0 0 0 0
24 27 1 0 0 0 0
28 27 1 0 0 0 0
29 28 1 0 0 0 0
23 29 1 0 0 0 0
29 30 1 6 0 0 0
28 31 1 6 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
36 38 2 0 0 0 0
38 39 1 0 0 0 0
38 40 1 0 0 0 0
40 41 2 0 0 0 0
31 41 1 0 0 0 0
34 41 1 0 0 0 0
33 42 1 0 0 0 0
42 43 1 0 0 0 0
44 43 1 6 0 0 0
44 45 1 0 0 0 0
46 45 1 0 0 0 0
47 46 1 0 0 0 0
47 48 1 1 0 0 0
47 49 1 0 0 0 0
49 44 1 0 0 0 0
49 50 1 1 0 0 0
46 51 1 6 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 54 1 0 0 0 0
54 55 2 0 0 0 0
55 56 1 0 0 0 0
55 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
51 60 1 0 0 0 0
54 60 1 0 0 0 0
42 61 1 0 0 0 0
2 61 1 0 0 0 0
61 62 1 0 0 0 0
8 62 1 0 0 0 0
62 63 2 0 0 0 0
63 64 1 0 0 0 0
63 65 1 0 0 0 0
65 66 2 0 0 0 0
42 66 1 0 0 0 0
66 67 1 0 0 0 0
67 68 2 0 0 0 0
68 69 1 0 0 0 0
69 70 2 0 0 0 0
42 70 1 0 0 0 0
70 71 1 0 0 0 0
71 72 2 0 0 0 0
72 73 1 0 0 0 0
72 74 1 0 0 0 0
74 75 2 0 0 0 0
42 75 1 0 0 0 0
76 75 1 0 0 0 0
2 76 1 0 0 0 0
76 77 1 1 0 0 0
76 78 1 0 0 0 0
78 79 1 1 0 0 0
78 80 1 6 0 0 0
80 81 1 0 0 0 0
81 82 1 0 0 0 0
81 83 2 0 0 0 0
78 84 1 0 0 0 0
84 74 1 0 0 0 0
84 85 1 1 0 0 0
85 86 1 0 0 0 0
86 87 1 0 0 0 0
87 88 1 0 0 0 0
87 89 2 0 0 0 0
90 71 1 0 0 0 0
90 91 1 1 0 0 0
90 92 1 6 0 0 0
92 93 1 0 0 0 0
93 94 1 0 0 0 0
93 95 2 0 0 0 0
90 96 1 0 0 0 0
96 69 1 0 0 0 0
96 97 1 1 0 0 0
97 98 1 0 0 0 0
98 99 1 0 0 0 0
99100 1 0 0 0 0
99101 2 0 0 0 0
67102 1 0 0 0 0
102103 1 0 0 0 0
102104 1 0 0 0 0
105102 1 0 0 0 0
105 65 1 0 0 0 0
105106 1 1 0 0 0
106107 1 0 0 0 0
107108 1 0 0 0 0
108109 1 0 0 0 0
108110 2 0 0 0 0
M CHG 6 20 -1 33 1 42 -3 66 1 70 1 75 1
M END
3D MOL for HMDB0002086 (Adenosylcobalamin)HMDB0002086
RDKit 3D
Adenosylcobalamin
209223 0 0 0 0 0 0 0 0999 V2000
6.0032 11.2820 0.2503 C 0 0 0 0 0 0 0 0 0 0 0 0
6.8948 10.1816 -0.1547 C 0 0 0 0 0 0 0 0 0 0 0 0
7.0401 9.1030 0.6780 C 0 0 0 0 0 0 0 0 0 0 0 0
7.4973 7.9876 0.2138 N 0 0 0 0 0 4 0 0 0 0 0 0
7.9035 7.1875 1.1113 C 0 0 0 0 0 0 0 0 0 0 0 0
8.9569 6.3454 0.9227 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5663 5.9539 -0.2227 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1175 6.1461 -1.4327 N 0 0 0 0 0 4 0 0 0 0 0 0
9.9150 5.6973 -2.3527 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5343 5.5506 -3.7253 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3780 4.7083 -4.5901 C 0 0 0 0 0 0 0 0 0 0 0 0
8.4388 6.1717 -4.2883 C 0 0 0 0 0 0 0 0 0 0 0 0
7.8399 7.1349 -3.6344 N 0 0 0 0 0 0 0 0 0 0 0 0
7.7385 8.3611 -4.3808 C 0 0 0 0 0 0 0 0 0 0 0 0
8.2609 7.7994 -5.7718 C 0 0 0 0 0 0 0 0 0 0 0 0
7.9570 8.6045 -7.0858 C 0 0 0 0 0 0 0 0 0 0 0 0
6.7909 8.2519 -7.9287 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9541 7.9156 -9.2247 N 0 0 0 0 0 0 0 0 0 0 0 0
5.6202 8.2938 -7.4934 O 0 0 0 0 0 0 0 0 0 0 0 0
7.8128 6.2678 -5.6436 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2259 6.0769 -5.5658 C 0 0 0 0 0 0 0 0 0 0 0 0
8.2274 5.2793 -6.8111 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7406 3.7784 -6.6850 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3437 3.4461 -7.0842 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0095 3.6105 -8.2814 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4539 2.9699 -6.1487 N 0 0 0 0 0 0 0 0 0 0 0 0
4.0950 2.5702 -6.3740 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0105 3.4181 -5.6263 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6087 3.0467 -6.2131 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1459 3.2482 -4.1759 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9910 3.8389 -3.2195 P 0 0 0 0 0 5 0 0 0 0 0 0
1.5381 5.1442 -3.7124 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7780 2.8205 -3.2675 O 0 0 0 0 0 1 0 0 0 0 0 0
2.5125 4.0390 -1.7154 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9827 3.2367 -0.6215 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8531 4.0807 0.6731 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8765 3.5054 1.7394 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9518 4.2600 2.9432 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1736 4.1759 1.2314 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9829 3.0989 0.7218 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0451 2.2304 -0.1753 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6207 1.5960 -1.3156 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1387 3.6058 0.0204 N 0 0 0 0 0 0 0 0 0 0 0 0
5.4497 4.8807 -0.3055 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4988 5.0037 -1.1529 N 0 0 0 0 0 4 0 0 0 0 0 0
6.8391 3.7027 -1.3471 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7444 3.0969 -2.2301 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0828 1.6884 -2.1651 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1269 1.0021 -3.0546 C 0 0 0 0 0 0 0 0 0 0 0 0
7.3876 0.8966 -1.1968 C 0 0 0 0 0 0 0 0 0 0 0 0
7.6525 -0.5609 -1.0250 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3972 1.4965 -0.3841 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1136 2.8668 -0.5150 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1935 7.1182 -1.6589 Co 0 0 0 0 0 6 0 0 0 0 0 0
4.9568 7.3883 -2.3284 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0039 8.2356 -1.4290 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4049 9.3209 -2.1700 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0285 9.0252 -2.4818 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8107 8.4936 -3.8117 N 0 0 0 0 0 0 0 0 0 0 0 0
2.7025 8.4167 -4.8281 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1735 7.8938 -5.9489 N 0 0 0 0 0 0 0 0 0 0 0 0
0.8930 7.6574 -5.6481 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1545 7.1251 -6.4165 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0327 6.7321 -7.6876 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.3797 7.0182 -5.8325 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.6047 7.4097 -4.5454 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5913 7.9253 -3.7960 N 0 0 0 0 0 0 0 0 0 0 0 0
0.6511 8.0431 -4.3329 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5457 8.1394 -1.3069 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8952 8.9491 -0.3231 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8262 7.4761 -0.7670 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8737 7.5093 0.6598 O 0 0 0 0 0 0 0 0 0 0 0 0
7.7680 9.2221 -2.1429 N 0 0 0 0 0 4 0 0 0 0 0 0
7.5337 10.2569 -1.4059 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0059 11.4773 -2.0793 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3120 12.1123 -1.4714 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1667 12.7815 -0.0766 C 0 0 0 0 0 0 0 0 0 0 0 0
10.4562 13.3269 0.4114 C 0 0 0 0 0 0 0 0 0 0 0 0
10.6353 14.6339 0.6791 N 0 0 0 0 0 0 0 0 0 0 0 0
11.4301 12.5616 0.5984 O 0 0 0 0 0 0 0 0 0 0 0 0
7.9748 11.0036 -3.5981 C 0 0 0 0 0 0 0 0 0 0 0 0
8.9664 11.8043 -4.5106 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4741 11.2828 -4.0966 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1125 11.2454 -5.5319 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5831 12.1363 -6.4241 N 0 0 0 0 0 0 0 0 0 0 0 0
5.2534 10.4375 -5.9429 O 0 0 0 0 0 0 0 0 0 0 0 0
8.3668 9.4627 -3.4196 C 0 0 0 0 0 0 0 0 0 0 0 0
9.9184 9.1905 -3.3739 C 0 0 0 0 0 0 0 0 0 0 0 0
11.2054 5.3140 -1.7295 C 0 0 0 0 0 0 0 0 0 0 0 0
12.4277 6.2612 -2.0595 C 0 0 0 0 0 0 0 0 0 0 0 0
13.0755 6.1638 -3.4689 C 0 0 0 0 0 0 0 0 0 0 0 0
14.4079 6.8027 -3.5725 C 0 0 0 0 0 0 0 0 0 0 0 0
14.5564 8.1357 -3.5001 N 0 0 0 0 0 0 0 0 0 0 0 0
15.4371 6.1121 -3.7547 O 0 0 0 0 0 0 0 0 0 0 0 0
10.8176 5.1970 -0.2074 C 0 0 0 0 0 0 0 0 0 0 0 0
11.8879 5.7610 0.8044 C 0 0 0 0 0 0 0 0 0 0 0 0
10.5249 3.7134 0.2169 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1428 7.4309 2.3211 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7449 6.9526 3.6937 C 0 0 0 0 0 0 0 0 0 0 0 0
7.5451 5.4375 3.9925 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0127 5.0645 5.3442 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3034 5.2001 5.6978 N 0 0 0 0 0 0 0 0 0 0 0 0
7.2118 4.6113 6.1931 O 0 0 0 0 0 0 0 0 0 0 0 0
6.8075 8.9634 2.1462 C 0 0 0 0 0 0 0 0 0 0 0 0
7.8604 9.8573 2.8967 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3461 9.2992 2.6453 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9192 8.7407 3.9511 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9950 9.4612 5.0867 N 0 0 0 0 0 0 0 0 0 0 0 0
4.4459 7.5835 4.0292 O 0 0 0 0 0 0 0 0 0 0 0 0
6.3866 11.8274 1.1091 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7946 12.0129 -0.5230 H 0 0 0 0 0 0 0 0 0 0 0 0
5.0175 10.8875 0.4854 H 0 0 0 0 0 0 0 0 0 0 0 0
9.3643 6.0208 1.7590 H 0 0 0 0 0 0 0 0 0 0 0 0
11.1667 4.1628 -4.0985 H 0 0 0 0 0 0 0 0 0 0 0 0
10.8578 5.3165 -5.3565 H 0 0 0 0 0 0 0 0 0 0 0 0
9.8261 3.9146 -5.0566 H 0 0 0 0 0 0 0 0 0 0 0 0
6.7055 8.6485 -4.5140 H 0 0 0 0 0 0 0 0 0 0 0 0
9.3631 7.8561 -5.7464 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8426 8.5308 -7.7293 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8876 9.6585 -6.8376 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8451 7.8874 -9.6124 H 0 0 0 0 0 0 0 0 0 0 0 0
6.1821 7.7101 -9.7750 H 0 0 0 0 0 0 0 0 0 0 0 0
5.9758 5.2377 -4.9136 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7684 5.9168 -6.5435 H 0 0 0 0 0 0 0 0 0 0 0 0
5.6799 6.9202 -5.1777 H 0 0 0 0 0 0 0 0 0 0 0 0
9.3039 5.2999 -6.9509 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8416 5.6691 -7.7543 H 0 0 0 0 0 0 0 0 0 0 0 0
8.4046 3.1528 -7.2885 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8421 3.4548 -5.6509 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7901 2.8641 -5.2452 H 0 0 0 0 0 0 0 0 0 0 0 0
3.8889 2.5628 -7.4482 H 0 0 0 0 0 0 0 0 0 0 0 0
4.0152 1.5359 -6.0370 H 0 0 0 0 0 0 0 0 0 0 0 0
3.1952 4.4723 -5.8642 H 0 0 0 0 0 0 0 0 0 0 0 0
1.3239 2.0340 -5.9176 H 0 0 0 0 0 0 0 0 0 0 0 0
1.6086 3.1094 -7.3025 H 0 0 0 0 0 0 0 0 0 0 0 0
0.8453 3.7401 -5.8621 H 0 0 0 0 0 0 0 0 0 0 0 0
1.0314 2.7437 -0.8409 H 0 0 0 0 0 0 0 0 0 0 0 0
1.4997 5.0847 0.4263 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.1496 3.5438 1.3660 H 0 0 0 0 0 0 0 0 0 0 0 0
1.1166 2.4702 1.9870 H 0 0 0 0 0 0 0 0 0 0 0 0
0.4040 3.7556 3.5807 H 0 0 0 0 0 0 0 0 0 0 0 0
4.3234 2.5106 1.5793 H 0 0 0 0 0 0 0 0 0 0 0 0
2.5846 1.4425 0.4276 H 0 0 0 0 0 0 0 0 0 0 0 0
4.1958 0.8893 -0.9610 H 0 0 0 0 0 0 0 0 0 0 0 0
4.9386 5.6850 0.0421 H 0 0 0 0 0 0 0 0 0 0 0 0
8.1680 3.6476 -2.9504 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9111 0.5546 -2.4416 H 0 0 0 0 0 0 0 0 0 0 0 0
8.6582 0.2319 -3.6692 H 0 0 0 0 0 0 0 0 0 0 0 0
9.5941 1.7289 -3.7190 H 0 0 0 0 0 0 0 0 0 0 0 0
7.4729 -1.0817 -1.9670 H 0 0 0 0 0 0 0 0 0 0 0 0
7.0132 -1.0237 -0.2697 H 0 0 0 0 0 0 0 0 0 0 0 0
8.6896 -0.7177 -0.7216 H 0 0 0 0 0 0 0 0 0 0 0 0
5.8722 0.9413 0.2950 H 0 0 0 0 0 0 0 0 0 0 0 0
5.0235 7.9064 -3.2658 H 0 0 0 0 0 0 0 0 0 0 0 0
4.5374 6.4080 -2.5574 H 0 0 0 0 0 0 0 0 0 0 0 0
4.6179 8.6842 -0.6572 H 0 0 0 0 0 0 0 0 0 0 0 0
1.4884 9.9769 -2.4622 H 0 0 0 0 0 0 0 0 0 0 0 0
3.6654 8.7359 -4.7861 H 0 0 0 0 0 0 0 0 0 0 0 0
0.9059 6.8200 -8.1014 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.7050 6.3593 -8.1946 H 0 0 0 0 0 0 0 0 0 0 0 0
-2.5364 7.3132 -4.1417 H 0 0 0 0 0 0 0 0 0 0 0 0
0.8313 7.3769 -1.6256 H 0 0 0 0 0 0 0 0 0 0 0 0
0.5641 8.3185 0.3510 H 0 0 0 0 0 0 0 0 0 0 0 0
2.8294 6.4324 -1.0642 H 0 0 0 0 0 0 0 0 0 0 0 0
3.7015 7.0484 0.9058 H 0 0 0 0 0 0 0 0 0 0 0 0
7.3065 12.3024 -1.9709 H 0 0 0 0 0 0 0 0 0 0 0 0
10.0983 11.3755 -1.3784 H 0 0 0 0 0 0 0 0 0 0 0 0
9.6685 12.8962 -2.1440 H 0 0 0 0 0 0 0 0 0 0 0 0
8.4098 13.5707 -0.1318 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8331 12.0402 0.6473 H 0 0 0 0 0 0 0 0 0 0 0 0
11.5030 14.9473 0.9865 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9058 15.2641 0.5654 H 0 0 0 0 0 0 0 0 0 0 0 0
8.6677 12.8522 -4.5771 H 0 0 0 0 0 0 0 0 0 0 0 0
8.9941 11.3766 -5.5124 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9875 11.7830 -4.1330 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7997 10.6091 -3.5591 H 0 0 0 0 0 0 0 0 0 0 0 0
6.1634 12.2917 -3.8069 H 0 0 0 0 0 0 0 0 0 0 0 0
7.1947 12.8335 -6.1415 H 0 0 0 0 0 0 0 0 0 0 0 0
6.3058 12.0853 -7.3535 H 0 0 0 0 0 0 0 0 0 0 0 0
10.4215 9.6956 -2.5683 H 0 0 0 0 0 0 0 0 0 0 0 0
10.1509 8.1495 -3.2254 H 0 0 0 0 0 0 0 0 0 0 0 0
10.3929 9.4803 -4.3111 H 0 0 0 0 0 0 0 0 0 0 0 0
11.5071 4.3109 -2.0250 H 0 0 0 0 0 0 0 0 0 0 0 0
13.2420 5.9931 -1.3882 H 0 0 0 0 0 0 0 0 0 0 0 0
12.1592 7.2987 -1.8489 H 0 0 0 0 0 0 0 0 0 0 0 0
13.2002 5.1104 -3.7250 H 0 0 0 0 0 0 0 0 0 0 0 0
12.4257 6.6323 -4.2058 H 0 0 0 0 0 0 0 0 0 0 0 0
15.4380 8.5305 -3.6003 H 0 0 0 0 0 0 0 0 0 0 0 0
13.7869 8.7067 -3.3583 H 0 0 0 0 0 0 0 0 0 0 0 0
11.5797 5.6394 1.8433 H 0 0 0 0 0 0 0 0 0 0 0 0
12.8325 5.2231 0.7042 H 0 0 0 0 0 0 0 0 0 0 0 0
12.0675 6.8252 0.6392 H 0 0 0 0 0 0 0 0 0 0 0 0
10.1554 3.6530 1.2438 H 0 0 0 0 0 0 0 0 0 0 0 0
11.4337 3.1112 0.1480 H 0 0 0 0 0 0 0 0 0 0 0 0
9.7801 3.2559 -0.4298 H 0 0 0 0 0 0 0 0 0 0 0 0
6.2111 6.8683 2.1789 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8077 7.2048 3.7393 H 0 0 0 0 0 0 0 0 0 0 0 0
7.2601 7.4953 4.5056 H 0 0 0 0 0 0 0 0 0 0 0 0
6.4820 5.1977 3.9024 H 0 0 0 0 0 0 0 0 0 0 0 0
8.0773 4.8251 3.2643 H 0 0 0 0 0 0 0 0 0 0 0 0
9.5900 4.9548 6.5909 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9489 5.5477 5.0635 H 0 0 0 0 0 0 0 0 0 0 0 0
7.7065 10.9162 2.7019 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8786 9.6063 2.5879 H 0 0 0 0 0 0 0 0 0 0 0 0
7.7946 9.7275 3.9776 H 0 0 0 0 0 0 0 0 0 0 0 0
5.2232 10.3807 2.7239 H 0 0 0 0 0 0 0 0 0 0 0 0
4.6253 8.9576 1.9032 H 0 0 0 0 0 0 0 0 0 0 0 0
4.6948 9.0759 5.9261 H 0 0 0 0 0 0 0 0 0 0 0 0
5.3452 10.3662 5.0694 H 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0
2 3 2 0
3 4 1 0
4 5 2 0
5 6 1 0
6 7 2 0
7 8 1 0
8 9 2 0
9 10 1 0
10 11 1 0
10 12 2 0
12 13 1 0
13 14 1 0
14 15 1 0
15 16 1 0
16 17 1 0
17 18 1 0
17 19 2 0
15 20 1 0
20 21 1 0
20 22 1 0
22 23 1 0
23 24 1 0
24 25 2 0
24 26 1 0
26 27 1 0
27 28 1 0
28 29 1 0
28 30 1 0
30 31 1 0
31 32 2 0
31 33 1 0
31 34 1 0
34 35 1 0
35 36 1 0
36 37 1 0
37 38 1 0
36 39 1 0
39 40 1 0
40 41 1 0
41 42 1 0
40 43 1 0
43 44 1 0
44 45 2 0
45 46 1 0
46 47 2 0
47 48 1 0
48 49 1 0
48 50 2 0
50 51 1 0
50 52 1 0
52 53 2 0
45 54 1 0
54 55 1 0
55 56 1 0
56 57 1 0
57 58 1 0
58 59 1 0
59 60 1 0
60 61 2 0
61 62 1 0
62 63 2 0
63 64 1 0
63 65 1 0
65 66 2 0
66 67 1 0
67 68 2 0
58 69 1 0
69 70 1 0
69 71 1 0
71 72 1 0
54 73 1 0
73 74 2 0
74 75 1 0
75 76 1 0
76 77 1 0
77 78 1 0
78 79 1 0
78 80 2 0
75 81 1 0
81 82 1 0
81 83 1 0
83 84 1 0
84 85 1 0
84 86 2 0
81 87 1 0
87 88 1 0
9 89 1 0
89 90 1 0
90 91 1 0
91 92 1 0
92 93 1 0
92 94 2 0
89 95 1 0
95 96 1 0
95 97 1 0
5 98 1 0
98 99 1 0
99100 1 0
100101 1 0
101102 1 0
101103 2 0
98104 1 0
104105 1 0
104106 1 0
106107 1 0
107108 1 0
107109 2 0
74 2 1 0
104 3 1 0
54 4 1 0
71 56 1 0
87 73 1 0
95 7 1 0
54 8 1 0
68 59 1 0
20 12 1 0
41 35 1 0
53 43 1 0
68 62 1 0
54 13 1 0
87 14 1 0
53 46 1 0
1110 1 0
1111 1 0
1112 1 0
6113 1 0
11114 1 0
11115 1 0
11116 1 0
14117 1 0
15118 1 0
16119 1 0
16120 1 0
18121 1 0
18122 1 0
21123 1 0
21124 1 0
21125 1 0
22126 1 0
22127 1 0
23128 1 0
23129 1 0
26130 1 0
27131 1 0
27132 1 0
28133 1 0
29134 1 0
29135 1 0
29136 1 0
35137 1 0
36138 1 0
37139 1 0
37140 1 0
38141 1 0
40142 1 0
41143 1 0
42144 1 0
44145 1 0
47146 1 0
49147 1 0
49148 1 0
49149 1 0
51150 1 0
51151 1 0
51152 1 0
52153 1 0
55154 1 0
55155 1 0
56156 1 0
58157 1 0
60158 1 0
64159 1 0
64160 1 0
66161 1 0
69162 1 0
70163 1 0
71164 1 0
72165 1 0
75166 1 0
76167 1 0
76168 1 0
77169 1 0
77170 1 0
79171 1 0
79172 1 0
82173 1 0
82174 1 0
82175 1 0
83176 1 0
83177 1 0
85178 1 0
85179 1 0
88180 1 0
88181 1 0
88182 1 0
89183 1 0
90184 1 0
90185 1 0
91186 1 0
91187 1 0
93188 1 0
93189 1 0
96190 1 0
96191 1 0
96192 1 0
97193 1 0
97194 1 0
97195 1 0
98196 1 0
99197 1 0
99198 1 0
100199 1 0
100200 1 0
102201 1 0
102202 1 0
105203 1 0
105204 1 0
105205 1 0
106206 1 0
106207 1 0
108208 1 0
108209 1 0
M CHG 6 4 1 8 1 33 -1 45 1 54 -3 73 1
M END
3D SDF for HMDB0002086 (Adenosylcobalamin)
Mrv1652305171818062D
110124 0 0 1 0 999 V2000
0.2847 3.6890 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.1654 2.9976 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.5048 2.5166 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.0023 3.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4991 3.9582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1479 4.7047 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.3212 3.8891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2545 1.7305 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.4032 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7433 1.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5281 0.9263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8944 1.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4840 2.9616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6968 1.8883 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.0455 2.7592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8655 2.8503 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.7212 2.3422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1966 3.6059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0166 3.6970 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
4.3840 4.3306 0.0000 O 0 5 0 0 0 0 0 0 0 0 0 0
4.7392 4.5486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3477 4.4526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1677 4.5436 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
6.5760 5.2605 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.3030 6.0918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5106 6.9125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.3839 5.0937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.4750 4.2737 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
6.7233 3.9338 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
6.4005 3.1540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.3072 3.8479 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
8.9436 4.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3838 3.6131 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
9.0909 2.8787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6393 3.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3798 4.2781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2019 4.6166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5718 4.4449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5697 5.3430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0234 3.8286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.2829 3.0455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2580 2.9728 0.0000 Co 0 7 0 0 0 0 0 0 0 0 0 0
0.4677 2.0601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2815 1.9756 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6884 1.2579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4967 1.4231 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.5893 2.2429 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.5360 2.3059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8383 2.5844 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.1530 3.4299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1054 0.8663 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.9137 1.0315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3206 0.3138 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.7637 -0.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8435 -1.1161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5945 -1.4576 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.1723 -1.5958 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
2.4213 -1.2543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3415 -0.4332 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
3.0127 0.0465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8300 2.5088 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.5705 1.7257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3198 0.9709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3332 0.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1257 0.7947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5424 1.5067 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
-3.3483 1.3305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0206 2.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1117 2.9745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3600 3.3145 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
-3.4510 4.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5612 4.7169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6536 5.5395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7362 4.7217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4767 3.9386 0.0000 N 0 3 0 0 0 0 0 0 0 0 0 0
-0.6517 3.9434 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.0525 4.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4013 4.7295 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.3381 5.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2795 5.4046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0067 6.0005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8926 6.0577 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
1.2271 6.8246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0716 5.2105 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.1957 6.0863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5085 6.9296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5568 7.8132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1436 8.3984 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-0.7983 8.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2590 4.3012 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-4.0664 5.1588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9595 4.9013 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4260 5.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3187 6.5417 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-6.2630 5.8135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6673 3.5843 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-5.4893 3.3133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1274 2.7045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.0020 2.6843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.4826 1.9987 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-7.5621 3.3516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4298 0.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9067 -0.2118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2789 0.6327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6741 0.1784 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.6523 -0.6852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8365 -1.5532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4025 -2.3163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5713 -2.4256 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-2.8229 -3.0752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2 1 1 6 0 0 0
2 3 1 0 0 0 0
3 4 1 6 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
5 7 2 0 0 0 0
3 8 1 0 0 0 0
8 9 1 6 0 0 0
8 10 1 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
12 14 1 0 0 0 0
14 15 1 0 0 0 0
16 15 1 0 0 0 0
16 17 1 6 0 0 0
16 18 1 0 0 0 0
18 19 1 0 0 0 0
19 20 1 0 0 0 0
19 21 2 0 0 0 0
19 22 1 0 0 0 0
23 22 1 6 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 0 0 0 0
24 27 1 0 0 0 0
28 27 1 0 0 0 0
29 28 1 0 0 0 0
23 29 1 0 0 0 0
29 30 1 6 0 0 0
28 31 1 6 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
36 38 2 0 0 0 0
38 39 1 0 0 0 0
38 40 1 0 0 0 0
40 41 2 0 0 0 0
31 41 1 0 0 0 0
34 41 1 0 0 0 0
33 42 1 0 0 0 0
42 43 1 0 0 0 0
44 43 1 6 0 0 0
44 45 1 0 0 0 0
46 45 1 0 0 0 0
47 46 1 0 0 0 0
47 48 1 1 0 0 0
47 49 1 0 0 0 0
49 44 1 0 0 0 0
49 50 1 1 0 0 0
46 51 1 6 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 54 1 0 0 0 0
54 55 2 0 0 0 0
55 56 1 0 0 0 0
55 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
51 60 1 0 0 0 0
54 60 1 0 0 0 0
42 61 1 0 0 0 0
2 61 1 0 0 0 0
61 62 1 0 0 0 0
8 62 1 0 0 0 0
62 63 2 0 0 0 0
63 64 1 0 0 0 0
63 65 1 0 0 0 0
65 66 2 0 0 0 0
42 66 1 0 0 0 0
66 67 1 0 0 0 0
67 68 2 0 0 0 0
68 69 1 0 0 0 0
69 70 2 0 0 0 0
42 70 1 0 0 0 0
70 71 1 0 0 0 0
71 72 2 0 0 0 0
72 73 1 0 0 0 0
72 74 1 0 0 0 0
74 75 2 0 0 0 0
42 75 1 0 0 0 0
76 75 1 0 0 0 0
2 76 1 0 0 0 0
76 77 1 1 0 0 0
76 78 1 0 0 0 0
78 79 1 1 0 0 0
78 80 1 6 0 0 0
80 81 1 0 0 0 0
81 82 1 0 0 0 0
81 83 2 0 0 0 0
78 84 1 0 0 0 0
84 74 1 0 0 0 0
84 85 1 1 0 0 0
85 86 1 0 0 0 0
86 87 1 0 0 0 0
87 88 1 0 0 0 0
87 89 2 0 0 0 0
90 71 1 0 0 0 0
90 91 1 1 0 0 0
90 92 1 6 0 0 0
92 93 1 0 0 0 0
93 94 1 0 0 0 0
93 95 2 0 0 0 0
90 96 1 0 0 0 0
96 69 1 0 0 0 0
96 97 1 1 0 0 0
97 98 1 0 0 0 0
98 99 1 0 0 0 0
99100 1 0 0 0 0
99101 2 0 0 0 0
67102 1 0 0 0 0
102103 1 0 0 0 0
102104 1 0 0 0 0
105102 1 0 0 0 0
105 65 1 0 0 0 0
105106 1 1 0 0 0
106107 1 0 0 0 0
107108 1 0 0 0 0
108109 1 0 0 0 0
108110 2 0 0 0 0
M CHG 6 20 -1 33 1 42 -3 66 1 70 1 75 1
M END
> <DATABASE_ID>
HMDB0002086
> <DATABASE_NAME>
hmdb
> <SMILES>
[C@H]1(C[Co-3]2345[N+]6=C7C(C)=C8N2[C@]([H])([C@H](CC(=O)N)[C@]8(CCC(=O)NC[C@@H](C)OP([O-])(=O)O[C@@H]2[C@@H](CO)O[C@H](N8C=[N+]3C3=CC(C)=C(C)C=C83)[C@@H]2O)C)[C@@]2(C)[C@@](CC(=O)N)([C@H](CCC(=O)N)C(C(C)=C3[N+]4=C(C=C6C([C@@H]7CCC(=O)N)(C)C)[C@@H](CCC(=O)N)[C@@]3(CC(=O)N)C)=[N+]52)C)O[C@@H](N2C3=NC=NC(=C3N=C2)N)[C@H](O)[C@@H]1O
> <INCHI_IDENTIFIER>
InChI=1S/C62H90N13O14P.C10H12N5O3.Co/c1-29-20-39-40(21-30(29)2)75(28-70-39)57-52(84)53(41(27-76)87-57)89-90(85,86)88-31(3)26-69-49(83)18-19-59(8)37(22-46(66)80)56-62(11)61(10,25-48(68)82)36(14-17-45(65)79)51(74-62)33(5)55-60(9,24-47(67)81)34(12-15-43(63)77)38(71-55)23-42-58(6,7)35(13-16-44(64)78)50(72-42)32(4)54(59)73-56;1-4-6(16)7(17)10(18-4)15-3-14-5-8(11)12-2-13-9(5)15;/h20-21,23,28,31,34-37,41,52-53,56-57,76,84H,12-19,22,24-27H2,1-11H3,(H15,63,64,65,66,67,68,69,71,72,73,74,77,78,79,80,81,82,83,85,86);2-4,6-7,10,16-17H,1H2,(H2,11,12,13);/q;;+2/p-2/t31-,34-,35-,36-,37+,41-,52-,53-,56-,57+,59-,60+,61+,62+;4-,6-,7-,10-;/m11./s1
> <INCHI_KEY>
ZIHHMGTYZOSFRC-OUCXYWSSSA-L
> <FORMULA>
C72H100CoN18O17P
> <MOLECULAR_WEIGHT>
1579.5818
> <EXACT_MASS>
1578.65834557
> <JCHEM_ACCEPTOR_COUNT>
20
> <JCHEM_ATOM_COUNT>
209
> <JCHEM_AVERAGE_POLARIZABILITY>
156.6699379932075
> <JCHEM_BIOAVAILABILITY>
0
> <JCHEM_DONOR_COUNT>
12
> <JCHEM_FORMAL_CHARGE>
0
> <JCHEM_GHOSE_FILTER>
0
> <JCHEM_IUPAC>
(10S,12R,13S,17R,23R,24R,25R,30S,35S,36S,40S,41S,42R,46R)-1-{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}-30,35,40-tris(2-carbamoylethyl)-24,36,41-tris(carbamoylmethyl)-46-hydroxy-12-(hydroxymethyl)-5,6,17,23,28,31,31,36,38,41,42-undecamethyl-15,20-dioxo-11,14,16-trioxa-2lambda5,9,19,26,43lambda5,44lambda5,45lambda5-heptaaza-15lambda5-phospha-1-cobaltadodecacyclo[27.14.1.1^{1,34}.1^{2,9}.1^{10,13}.0^{1,26}.0^{3,8}.0^{23,27}.0^{25,42}.0^{32,44}.0^{39,43}.0^{37,45}]heptatetraconta-2(47),3,5,7,27,29(44),32,34(45),37,39(43)-decaene-2,43,44,45-tetrakis(ylium)-1,1,1-triuid-15-olate
> <JCHEM_LOGP>
-14.52923067663249
> <JCHEM_MDDR_LIKE_RULE>
1
> <JCHEM_NUMBER_OF_RINGS>
15
> <JCHEM_PHYSIOLOGICAL_CHARGE>
0
> <JCHEM_PKA>
12.228141104743376
> <JCHEM_PKA_STRONGEST_ACIDIC>
1.813329045543417
> <JCHEM_PKA_STRONGEST_BASIC>
3.9247140846726385
> <JCHEM_POLAR_SURFACE_AREA>
536.3099999999998
> <JCHEM_REFRACTIVITY>
397.8517
> <JCHEM_ROTATABLE_BOND_COUNT>
19
> <JCHEM_RULE_OF_FIVE>
0
> <JCHEM_TRADITIONAL_IUPAC>
(10S,12R,13S,17R,23R,24R,25R,30S,35S,36S,40S,41S,42R,46R)-1-{[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}-30,35,40-tris(2-carbamoylethyl)-24,36,41-tris(carbamoylmethyl)-46-hydroxy-12-(hydroxymethyl)-5,6,17,23,28,31,31,36,38,41,42-undecamethyl-15,20-dioxo-11,14,16-trioxa-2lambda5,9,19,26,43lambda5,44lambda5,45lambda5-heptaaza-15lambda5-phospha-1-cobaltadodecacyclo[27.14.1.1^{1,34}.1^{2,9}.1^{10,13}.0^{1,26}.0^{3,8}.0^{23,27}.0^{25,42}.0^{32,44}.0^{39,43}.0^{37,45}]heptatetraconta-2(47),3,5,7,27,29(44),32,34(45),37,39(43)-decaene-2,43,44,45-tetrakis(ylium)-1,1,1-triuid-15-olate
> <JCHEM_VEBER_RULE>
0
$$$$
3D-SDF for HMDB0002086 (Adenosylcobalamin)HMDB0002086
RDKit 3D
Adenosylcobalamin
209223 0 0 0 0 0 0 0 0999 V2000
6.0032 11.2820 0.2503 C 0 0 0 0 0 0 0 0 0 0 0 0
6.8948 10.1816 -0.1547 C 0 0 0 0 0 0 0 0 0 0 0 0
7.0401 9.1030 0.6780 C 0 0 0 0 0 0 0 0 0 0 0 0
7.4973 7.9876 0.2138 N 0 0 0 0 0 4 0 0 0 0 0 0
7.9035 7.1875 1.1113 C 0 0 0 0 0 0 0 0 0 0 0 0
8.9569 6.3454 0.9227 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5663 5.9539 -0.2227 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1175 6.1461 -1.4327 N 0 0 0 0 0 4 0 0 0 0 0 0
9.9150 5.6973 -2.3527 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5343 5.5506 -3.7253 C 0 0 0 0 0 0 0 0 0 0 0 0
10.3780 4.7083 -4.5901 C 0 0 0 0 0 0 0 0 0 0 0 0
8.4388 6.1717 -4.2883 C 0 0 0 0 0 0 0 0 0 0 0 0
7.8399 7.1349 -3.6344 N 0 0 0 0 0 0 0 0 0 0 0 0
7.7385 8.3611 -4.3808 C 0 0 0 0 0 0 0 0 0 0 0 0
8.2609 7.7994 -5.7718 C 0 0 0 0 0 0 0 0 0 0 0 0
7.9570 8.6045 -7.0858 C 0 0 0 0 0 0 0 0 0 0 0 0
6.7909 8.2519 -7.9287 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9541 7.9156 -9.2247 N 0 0 0 0 0 0 0 0 0 0 0 0
5.6202 8.2938 -7.4934 O 0 0 0 0 0 0 0 0 0 0 0 0
7.8128 6.2678 -5.6436 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2259 6.0769 -5.5658 C 0 0 0 0 0 0 0 0 0 0 0 0
8.2274 5.2793 -6.8111 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7406 3.7784 -6.6850 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3437 3.4461 -7.0842 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0095 3.6105 -8.2814 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4539 2.9699 -6.1487 N 0 0 0 0 0 0 0 0 0 0 0 0
4.0950 2.5702 -6.3740 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0105 3.4181 -5.6263 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6087 3.0467 -6.2131 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1459 3.2482 -4.1759 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9910 3.8389 -3.2195 P 0 0 0 0 0 5 0 0 0 0 0 0
1.5381 5.1442 -3.7124 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7780 2.8205 -3.2675 O 0 0 0 0 0 1 0 0 0 0 0 0
2.5125 4.0390 -1.7154 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9827 3.2367 -0.6215 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8531 4.0807 0.6731 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8765 3.5054 1.7394 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9518 4.2600 2.9432 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1736 4.1759 1.2314 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9829 3.0989 0.7218 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0451 2.2304 -0.1753 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6207 1.5960 -1.3156 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1387 3.6058 0.0204 N 0 0 0 0 0 0 0 0 0 0 0 0
5.4497 4.8807 -0.3055 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4988 5.0037 -1.1529 N 0 0 0 0 0 4 0 0 0 0 0 0
6.8391 3.7027 -1.3471 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7444 3.0969 -2.2301 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0828 1.6884 -2.1651 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1269 1.0021 -3.0546 C 0 0 0 0 0 0 0 0 0 0 0 0
7.3876 0.8966 -1.1968 C 0 0 0 0 0 0 0 0 0 0 0 0
7.6525 -0.5609 -1.0250 C 0 0 0 0 0 0 0 0 0 0 0 0
6.3972 1.4965 -0.3841 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1136 2.8668 -0.5150 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1935 7.1182 -1.6589 Co 0 0 0 0 0 6 0 0 0 0 0 0
4.9568 7.3883 -2.3284 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0039 8.2356 -1.4290 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4049 9.3209 -2.1700 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0285 9.0252 -2.4818 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8107 8.4936 -3.8117 N 0 0 0 0 0 0 0 0 0 0 0 0
2.7025 8.4167 -4.8281 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1735 7.8938 -5.9489 N 0 0 0 0 0 0 0 0 0 0 0 0
0.8930 7.6574 -5.6481 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1545 7.1251 -6.4165 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0327 6.7321 -7.6876 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.3797 7.0182 -5.8325 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.6047 7.4097 -4.5454 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5913 7.9253 -3.7960 N 0 0 0 0 0 0 0 0 0 0 0 0
0.6511 8.0431 -4.3329 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5457 8.1394 -1.3069 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8952 8.9491 -0.3231 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8262 7.4761 -0.7670 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8737 7.5093 0.6598 O 0 0 0 0 0 0 0 0 0 0 0 0
7.7680 9.2221 -2.1429 N 0 0 0 0 0 4 0 0 0 0 0 0
7.5337 10.2569 -1.4059 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0059 11.4773 -2.0793 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3120 12.1123 -1.4714 C 0 0 0 0 0 0 0 0 0 0 0 0
9.1667 12.7815 -0.0766 C 0 0 0 0 0 0 0 0 0 0 0 0
10.4562 13.3269 0.4114 C 0 0 0 0 0 0 0 0 0 0 0 0
10.6353 14.6339 0.6791 N 0 0 0 0 0 0 0 0 0 0 0 0
11.4301 12.5616 0.5984 O 0 0 0 0 0 0 0 0 0 0 0 0
7.9748 11.0036 -3.5981 C 0 0 0 0 0 0 0 0 0 0 0 0
8.9664 11.8043 -4.5106 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4741 11.2828 -4.0966 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1125 11.2454 -5.5319 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5831 12.1363 -6.4241 N 0 0 0 0 0 0 0 0 0 0 0 0
5.2534 10.4375 -5.9429 O 0 0 0 0 0 0 0 0 0 0 0 0
8.3668 9.4627 -3.4196 C 0 0 0 0 0 0 0 0 0 0 0 0
9.9184 9.1905 -3.3739 C 0 0 0 0 0 0 0 0 0 0 0 0
11.2054 5.3140 -1.7295 C 0 0 0 0 0 0 0 0 0 0 0 0
12.4277 6.2612 -2.0595 C 0 0 0 0 0 0 0 0 0 0 0 0
13.0755 6.1638 -3.4689 C 0 0 0 0 0 0 0 0 0 0 0 0
14.4079 6.8027 -3.5725 C 0 0 0 0 0 0 0 0 0 0 0 0
14.5564 8.1357 -3.5001 N 0 0 0 0 0 0 0 0 0 0 0 0
15.4371 6.1121 -3.7547 O 0 0 0 0 0 0 0 0 0 0 0 0
10.8176 5.1970 -0.2074 C 0 0 0 0 0 0 0 0 0 0 0 0
11.8879 5.7610 0.8044 C 0 0 0 0 0 0 0 0 0 0 0 0
10.5249 3.7134 0.2169 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1428 7.4309 2.3211 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7449 6.9526 3.6937 C 0 0 0 0 0 0 0 0 0 0 0 0
7.5451 5.4375 3.9925 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0127 5.0645 5.3442 C 0 0 0 0 0 0 0 0 0 0 0 0
9.3034 5.2001 5.6978 N 0 0 0 0 0 0 0 0 0 0 0 0
7.2118 4.6113 6.1931 O 0 0 0 0 0 0 0 0 0 0 0 0
6.8075 8.9634 2.1462 C 0 0 0 0 0 0 0 0 0 0 0 0
7.8604 9.8573 2.8967 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3461 9.2992 2.6453 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9192 8.7407 3.9511 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9950 9.4612 5.0867 N 0 0 0 0 0 0 0 0 0 0 0 0
4.4459 7.5835 4.0292 O 0 0 0 0 0 0 0 0 0 0 0 0
6.3866 11.8274 1.1091 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7946 12.0129 -0.5230 H 0 0 0 0 0 0 0 0 0 0 0 0
5.0175 10.8875 0.4854 H 0 0 0 0 0 0 0 0 0 0 0 0
9.3643 6.0208 1.7590 H 0 0 0 0 0 0 0 0 0 0 0 0
11.1667 4.1628 -4.0985 H 0 0 0 0 0 0 0 0 0 0 0 0
10.8578 5.3165 -5.3565 H 0 0 0 0 0 0 0 0 0 0 0 0
9.8261 3.9146 -5.0566 H 0 0 0 0 0 0 0 0 0 0 0 0
6.7055 8.6485 -4.5140 H 0 0 0 0 0 0 0 0 0 0 0 0
9.3631 7.8561 -5.7464 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8426 8.5308 -7.7293 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8876 9.6585 -6.8376 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8451 7.8874 -9.6124 H 0 0 0 0 0 0 0 0 0 0 0 0
6.1821 7.7101 -9.7750 H 0 0 0 0 0 0 0 0 0 0 0 0
5.9758 5.2377 -4.9136 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7684 5.9168 -6.5435 H 0 0 0 0 0 0 0 0 0 0 0 0
5.6799 6.9202 -5.1777 H 0 0 0 0 0 0 0 0 0 0 0 0
9.3039 5.2999 -6.9509 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8416 5.6691 -7.7543 H 0 0 0 0 0 0 0 0 0 0 0 0
8.4046 3.1528 -7.2885 H 0 0 0 0 0 0 0 0 0 0 0 0
7.8421 3.4548 -5.6509 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7901 2.8641 -5.2452 H 0 0 0 0 0 0 0 0 0 0 0 0
3.8889 2.5628 -7.4482 H 0 0 0 0 0 0 0 0 0 0 0 0
4.0152 1.5359 -6.0370 H 0 0 0 0 0 0 0 0 0 0 0 0
3.1952 4.4723 -5.8642 H 0 0 0 0 0 0 0 0 0 0 0 0
1.3239 2.0340 -5.9176 H 0 0 0 0 0 0 0 0 0 0 0 0
1.6086 3.1094 -7.3025 H 0 0 0 0 0 0 0 0 0 0 0 0
0.8453 3.7401 -5.8621 H 0 0 0 0 0 0 0 0 0 0 0 0
1.0314 2.7437 -0.8409 H 0 0 0 0 0 0 0 0 0 0 0 0
1.4997 5.0847 0.4263 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.1496 3.5438 1.3660 H 0 0 0 0 0 0 0 0 0 0 0 0
1.1166 2.4702 1.9870 H 0 0 0 0 0 0 0 0 0 0 0 0
0.4040 3.7556 3.5807 H 0 0 0 0 0 0 0 0 0 0 0 0
4.3234 2.5106 1.5793 H 0 0 0 0 0 0 0 0 0 0 0 0
2.5846 1.4425 0.4276 H 0 0 0 0 0 0 0 0 0 0 0 0
4.1958 0.8893 -0.9610 H 0 0 0 0 0 0 0 0 0 0 0 0
4.9386 5.6850 0.0421 H 0 0 0 0 0 0 0 0 0 0 0 0
8.1680 3.6476 -2.9504 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9111 0.5546 -2.4416 H 0 0 0 0 0 0 0 0 0 0 0 0
8.6582 0.2319 -3.6692 H 0 0 0 0 0 0 0 0 0 0 0 0
9.5941 1.7289 -3.7190 H 0 0 0 0 0 0 0 0 0 0 0 0
7.4729 -1.0817 -1.9670 H 0 0 0 0 0 0 0 0 0 0 0 0
7.0132 -1.0237 -0.2697 H 0 0 0 0 0 0 0 0 0 0 0 0
8.6896 -0.7177 -0.7216 H 0 0 0 0 0 0 0 0 0 0 0 0
5.8722 0.9413 0.2950 H 0 0 0 0 0 0 0 0 0 0 0 0
5.0235 7.9064 -3.2658 H 0 0 0 0 0 0 0 0 0 0 0 0
4.5374 6.4080 -2.5574 H 0 0 0 0 0 0 0 0 0 0 0 0
4.6179 8.6842 -0.6572 H 0 0 0 0 0 0 0 0 0 0 0 0
1.4884 9.9769 -2.4622 H 0 0 0 0 0 0 0 0 0 0 0 0
3.6654 8.7359 -4.7861 H 0 0 0 0 0 0 0 0 0 0 0 0
0.9059 6.8200 -8.1014 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.7050 6.3593 -8.1946 H 0 0 0 0 0 0 0 0 0 0 0 0
-2.5364 7.3132 -4.1417 H 0 0 0 0 0 0 0 0 0 0 0 0
0.8313 7.3769 -1.6256 H 0 0 0 0 0 0 0 0 0 0 0 0
0.5641 8.3185 0.3510 H 0 0 0 0 0 0 0 0 0 0 0 0
2.8294 6.4324 -1.0642 H 0 0 0 0 0 0 0 0 0 0 0 0
3.7015 7.0484 0.9058 H 0 0 0 0 0 0 0 0 0 0 0 0
7.3065 12.3024 -1.9709 H 0 0 0 0 0 0 0 0 0 0 0 0
10.0983 11.3755 -1.3784 H 0 0 0 0 0 0 0 0 0 0 0 0
9.6685 12.8962 -2.1440 H 0 0 0 0 0 0 0 0 0 0 0 0
8.4098 13.5707 -0.1318 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8331 12.0402 0.6473 H 0 0 0 0 0 0 0 0 0 0 0 0
11.5030 14.9473 0.9865 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9058 15.2641 0.5654 H 0 0 0 0 0 0 0 0 0 0 0 0
8.6677 12.8522 -4.5771 H 0 0 0 0 0 0 0 0 0 0 0 0
8.9941 11.3766 -5.5124 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9875 11.7830 -4.1330 H 0 0 0 0 0 0 0 0 0 0 0 0
5.7997 10.6091 -3.5591 H 0 0 0 0 0 0 0 0 0 0 0 0
6.1634 12.2917 -3.8069 H 0 0 0 0 0 0 0 0 0 0 0 0
7.1947 12.8335 -6.1415 H 0 0 0 0 0 0 0 0 0 0 0 0
6.3058 12.0853 -7.3535 H 0 0 0 0 0 0 0 0 0 0 0 0
10.4215 9.6956 -2.5683 H 0 0 0 0 0 0 0 0 0 0 0 0
10.1509 8.1495 -3.2254 H 0 0 0 0 0 0 0 0 0 0 0 0
10.3929 9.4803 -4.3111 H 0 0 0 0 0 0 0 0 0 0 0 0
11.5071 4.3109 -2.0250 H 0 0 0 0 0 0 0 0 0 0 0 0
13.2420 5.9931 -1.3882 H 0 0 0 0 0 0 0 0 0 0 0 0
12.1592 7.2987 -1.8489 H 0 0 0 0 0 0 0 0 0 0 0 0
13.2002 5.1104 -3.7250 H 0 0 0 0 0 0 0 0 0 0 0 0
12.4257 6.6323 -4.2058 H 0 0 0 0 0 0 0 0 0 0 0 0
15.4380 8.5305 -3.6003 H 0 0 0 0 0 0 0 0 0 0 0 0
13.7869 8.7067 -3.3583 H 0 0 0 0 0 0 0 0 0 0 0 0
11.5797 5.6394 1.8433 H 0 0 0 0 0 0 0 0 0 0 0 0
12.8325 5.2231 0.7042 H 0 0 0 0 0 0 0 0 0 0 0 0
12.0675 6.8252 0.6392 H 0 0 0 0 0 0 0 0 0 0 0 0
10.1554 3.6530 1.2438 H 0 0 0 0 0 0 0 0 0 0 0 0
11.4337 3.1112 0.1480 H 0 0 0 0 0 0 0 0 0 0 0 0
9.7801 3.2559 -0.4298 H 0 0 0 0 0 0 0 0 0 0 0 0
6.2111 6.8683 2.1789 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8077 7.2048 3.7393 H 0 0 0 0 0 0 0 0 0 0 0 0
7.2601 7.4953 4.5056 H 0 0 0 0 0 0 0 0 0 0 0 0
6.4820 5.1977 3.9024 H 0 0 0 0 0 0 0 0 0 0 0 0
8.0773 4.8251 3.2643 H 0 0 0 0 0 0 0 0 0 0 0 0
9.5900 4.9548 6.5909 H 0 0 0 0 0 0 0 0 0 0 0 0
9.9489 5.5477 5.0635 H 0 0 0 0 0 0 0 0 0 0 0 0
7.7065 10.9162 2.7019 H 0 0 0 0 0 0 0 0 0 0 0 0
8.8786 9.6063 2.5879 H 0 0 0 0 0 0 0 0 0 0 0 0
7.7946 9.7275 3.9776 H 0 0 0 0 0 0 0 0 0 0 0 0
5.2232 10.3807 2.7239 H 0 0 0 0 0 0 0 0 0 0 0 0
4.6253 8.9576 1.9032 H 0 0 0 0 0 0 0 0 0 0 0 0
4.6948 9.0759 5.9261 H 0 0 0 0 0 0 0 0 0 0 0 0
5.3452 10.3662 5.0694 H 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0
2 3 2 0
3 4 1 0
4 5 2 0
5 6 1 0
6 7 2 0
7 8 1 0
8 9 2 0
9 10 1 0
10 11 1 0
10 12 2 0
12 13 1 0
13 14 1 0
14 15 1 0
15 16 1 0
16 17 1 0
17 18 1 0
17 19 2 0
15 20 1 0
20 21 1 0
20 22 1 0
22 23 1 0
23 24 1 0
24 25 2 0
24 26 1 0
26 27 1 0
27 28 1 0
28 29 1 0
28 30 1 0
30 31 1 0
31 32 2 0
31 33 1 0
31 34 1 0
34 35 1 0
35 36 1 0
36 37 1 0
37 38 1 0
36 39 1 0
39 40 1 0
40 41 1 0
41 42 1 0
40 43 1 0
43 44 1 0
44 45 2 0
45 46 1 0
46 47 2 0
47 48 1 0
48 49 1 0
48 50 2 0
50 51 1 0
50 52 1 0
52 53 2 0
45 54 1 0
54 55 1 0
55 56 1 0
56 57 1 0
57 58 1 0
58 59 1 0
59 60 1 0
60 61 2 0
61 62 1 0
62 63 2 0
63 64 1 0
63 65 1 0
65 66 2 0
66 67 1 0
67 68 2 0
58 69 1 0
69 70 1 0
69 71 1 0
71 72 1 0
54 73 1 0
73 74 2 0
74 75 1 0
75 76 1 0
76 77 1 0
77 78 1 0
78 79 1 0
78 80 2 0
75 81 1 0
81 82 1 0
81 83 1 0
83 84 1 0
84 85 1 0
84 86 2 0
81 87 1 0
87 88 1 0
9 89 1 0
89 90 1 0
90 91 1 0
91 92 1 0
92 93 1 0
92 94 2 0
89 95 1 0
95 96 1 0
95 97 1 0
5 98 1 0
98 99 1 0
99100 1 0
100101 1 0
101102 1 0
101103 2 0
98104 1 0
104105 1 0
104106 1 0
106107 1 0
107108 1 0
107109 2 0
74 2 1 0
104 3 1 0
54 4 1 0
71 56 1 0
87 73 1 0
95 7 1 0
54 8 1 0
68 59 1 0
20 12 1 0
41 35 1 0
53 43 1 0
68 62 1 0
54 13 1 0
87 14 1 0
53 46 1 0
1110 1 0
1111 1 0
1112 1 0
6113 1 0
11114 1 0
11115 1 0
11116 1 0
14117 1 0
15118 1 0
16119 1 0
16120 1 0
18121 1 0
18122 1 0
21123 1 0
21124 1 0
21125 1 0
22126 1 0
22127 1 0
23128 1 0
23129 1 0
26130 1 0
27131 1 0
27132 1 0
28133 1 0
29134 1 0
29135 1 0
29136 1 0
35137 1 0
36138 1 0
37139 1 0
37140 1 0
38141 1 0
40142 1 0
41143 1 0
42144 1 0
44145 1 0
47146 1 0
49147 1 0
49148 1 0
49149 1 0
51150 1 0
51151 1 0
51152 1 0
52153 1 0
55154 1 0
55155 1 0
56156 1 0
58157 1 0
60158 1 0
64159 1 0
64160 1 0
66161 1 0
69162 1 0
70163 1 0
71164 1 0
72165 1 0
75166 1 0
76167 1 0
76168 1 0
77169 1 0
77170 1 0
79171 1 0
79172 1 0
82173 1 0
82174 1 0
82175 1 0
83176 1 0
83177 1 0
85178 1 0
85179 1 0
88180 1 0
88181 1 0
88182 1 0
89183 1 0
90184 1 0
90185 1 0
91186 1 0
91187 1 0
93188 1 0
93189 1 0
96190 1 0
96191 1 0
96192 1 0
97193 1 0
97194 1 0
97195 1 0
98196 1 0
99197 1 0
99198 1 0
100199 1 0
100200 1 0
102201 1 0
102202 1 0
105203 1 0
105204 1 0
105205 1 0
106206 1 0
106207 1 0
108208 1 0
108209 1 0
M CHG 6 4 1 8 1 33 -1 45 1 54 -3 73 1
M END
PDB for HMDB0002086 (Adenosylcobalamin)HEADER PROTEIN 17-MAY-18 NONE TITLE NULL COMPND NULL SOURCE NULL KEYWDS NULL EXPDTA NULL AUTHOR Marvin REVDAT 1 17-MAY-18 0 HETATM 1 H UNK 0 0.531 6.886 0.000 0.00 0.00 H+0 HETATM 2 C UNK 0 -0.309 5.596 0.000 0.00 0.00 C+0 HETATM 3 C UNK 0 0.942 4.698 0.000 0.00 0.00 C+0 HETATM 4 C UNK 0 1.871 6.055 0.000 0.00 0.00 C+0 HETATM 5 C UNK 0 2.798 7.389 0.000 0.00 0.00 C+0 HETATM 6 N UNK 0 2.143 8.782 0.000 0.00 0.00 N+0 HETATM 7 O UNK 0 4.333 7.260 0.000 0.00 0.00 O+0 HETATM 8 C UNK 0 0.475 3.230 0.000 0.00 0.00 C+0 HETATM 9 C UNK 0 -0.753 1.828 0.000 0.00 0.00 C+0 HETATM 10 C UNK 0 1.387 1.990 0.000 0.00 0.00 C+0 HETATM 11 C UNK 0 2.852 1.729 0.000 0.00 0.00 C+0 HETATM 12 C UNK 0 3.536 3.570 0.000 0.00 0.00 C+0 HETATM 13 O UNK 0 2.770 5.528 0.000 0.00 0.00 O+0 HETATM 14 N UNK 0 5.034 3.525 0.000 0.00 0.00 N+0 HETATM 15 C UNK 0 5.685 5.151 0.000 0.00 0.00 C+0 HETATM 16 C UNK 0 7.216 5.321 0.000 0.00 0.00 C+0 HETATM 17 C UNK 0 8.813 4.372 0.000 0.00 0.00 C+0 HETATM 18 O UNK 0 7.834 6.731 0.000 0.00 0.00 O+0 HETATM 19 P UNK 0 9.364 6.901 0.000 0.00 0.00 P+0 HETATM 20 O UNK 0 8.183 8.084 0.000 0.00 0.00 O-1 HETATM 21 O UNK 0 8.847 8.491 0.000 0.00 0.00 O+0 HETATM 22 O UNK 0 9.982 8.312 0.000 0.00 0.00 O+0 HETATM 23 C UNK 0 11.513 8.481 0.000 0.00 0.00 C+0 HETATM 24 C UNK 0 12.275 9.820 0.000 0.00 0.00 C+0 HETATM 25 C UNK 0 11.766 11.371 0.000 0.00 0.00 C+0 HETATM 26 O UNK 0 12.153 12.903 0.000 0.00 0.00 O+0 HETATM 27 O UNK 0 13.783 9.508 0.000 0.00 0.00 O+0 HETATM 28 C UNK 0 13.953 7.978 0.000 0.00 0.00 C+0 HETATM 29 C UNK 0 12.550 7.343 0.000 0.00 0.00 C+0 HETATM 30 O UNK 0 11.948 5.887 0.000 0.00 0.00 O+0 HETATM 31 N UNK 0 15.507 7.183 0.000 0.00 0.00 N+0 HETATM 32 C UNK 0 16.695 7.850 0.000 0.00 0.00 C+0 HETATM 33 N UNK 0 17.516 6.744 0.000 0.00 0.00 N+1 HETATM 34 C UNK 0 16.970 5.374 0.000 0.00 0.00 C+0 HETATM 35 C UNK 0 17.993 6.524 0.000 0.00 0.00 C+0 HETATM 36 C UNK 0 17.509 7.986 0.000 0.00 0.00 C+0 HETATM 37 C UNK 0 19.044 8.618 0.000 0.00 0.00 C+0 HETATM 38 C UNK 0 16.001 8.297 0.000 0.00 0.00 C+0 HETATM 39 C UNK 0 15.997 9.974 0.000 0.00 0.00 C+0 HETATM 40 C UNK 0 14.977 7.147 0.000 0.00 0.00 C+0 HETATM 41 C UNK 0 15.461 5.685 0.000 0.00 0.00 C+0 HETATM 42 Co UNK 0 0.482 5.549 0.000 0.00 0.00 Co-3 HETATM 43 C UNK 0 0.873 3.846 0.000 0.00 0.00 C+0 HETATM 44 C UNK 0 2.392 3.688 0.000 0.00 0.00 C+0 HETATM 45 O UNK 0 3.152 2.348 0.000 0.00 0.00 O+0 HETATM 46 C UNK 0 4.661 2.656 0.000 0.00 0.00 C+0 HETATM 47 C UNK 0 4.833 4.187 0.000 0.00 0.00 C+0 HETATM 48 O UNK 0 6.601 4.304 0.000 0.00 0.00 O+0 HETATM 49 C UNK 0 3.431 4.824 0.000 0.00 0.00 C+0 HETATM 50 O UNK 0 4.019 6.402 0.000 0.00 0.00 O+0 HETATM 51 N UNK 0 5.797 1.617 0.000 0.00 0.00 N+0 HETATM 52 C UNK 0 7.306 1.925 0.000 0.00 0.00 C+0 HETATM 53 N UNK 0 8.065 0.586 0.000 0.00 0.00 N+0 HETATM 54 C UNK 0 7.026 -0.551 0.000 0.00 0.00 C+0 HETATM 55 C UNK 0 7.175 -2.083 0.000 0.00 0.00 C+0 HETATM 56 N UNK 0 8.576 -2.721 0.000 0.00 0.00 N+0 HETATM 57 N UNK 0 5.922 -2.979 0.000 0.00 0.00 N+0 HETATM 58 C UNK 0 4.520 -2.341 0.000 0.00 0.00 C+0 HETATM 59 N UNK 0 4.371 -0.809 0.000 0.00 0.00 N+0 HETATM 60 C UNK 0 5.624 0.087 0.000 0.00 0.00 C+0 HETATM 61 N UNK 0 -1.549 4.683 0.000 0.00 0.00 N+0 HETATM 62 C UNK 0 -1.065 3.221 0.000 0.00 0.00 C+0 HETATM 63 C UNK 0 -2.464 1.812 0.000 0.00 0.00 C+0 HETATM 64 C UNK 0 -2.489 0.183 0.000 0.00 0.00 C+0 HETATM 65 C UNK 0 -3.968 1.483 0.000 0.00 0.00 C+0 HETATM 66 N UNK 0 -4.746 2.813 0.000 0.00 0.00 N+1 HETATM 67 C UNK 0 -6.250 2.484 0.000 0.00 0.00 C+0 HETATM 68 C UNK 0 -7.505 4.022 0.000 0.00 0.00 C+0 HETATM 69 C UNK 0 -7.675 5.552 0.000 0.00 0.00 C+0 HETATM 70 N UNK 0 -6.272 6.187 0.000 0.00 0.00 N+1 HETATM 71 C UNK 0 -6.442 7.718 0.000 0.00 0.00 C+0 HETATM 72 C UNK 0 -4.781 8.805 0.000 0.00 0.00 C+0 HETATM 73 C UNK 0 -4.953 10.340 0.000 0.00 0.00 C+0 HETATM 74 C UNK 0 -3.241 8.814 0.000 0.00 0.00 C+0 HETATM 75 N UNK 0 -2.757 7.352 0.000 0.00 0.00 N+1 HETATM 76 C UNK 0 -1.217 7.361 0.000 0.00 0.00 C+0 HETATM 77 C UNK 0 0.098 8.114 0.000 0.00 0.00 C+0 HETATM 78 C UNK 0 -0.749 8.828 0.000 0.00 0.00 C+0 HETATM 79 C UNK 0 0.631 9.487 0.000 0.00 0.00 C+0 HETATM 80 C UNK 0 0.522 10.089 0.000 0.00 0.00 C+0 HETATM 81 C UNK 0 1.879 11.201 0.000 0.00 0.00 C+0 HETATM 82 N UNK 0 3.533 11.308 0.000 0.00 0.00 N+0 HETATM 83 O UNK 0 2.291 12.739 0.000 0.00 0.00 O+0 HETATM 84 C UNK 0 -2.000 9.726 0.000 0.00 0.00 C+0 HETATM 85 C UNK 0 -2.232 11.361 0.000 0.00 0.00 C+0 HETATM 86 C UNK 0 -2.816 12.935 0.000 0.00 0.00 C+0 HETATM 87 C UNK 0 -2.906 14.585 0.000 0.00 0.00 C+0 HETATM 88 N UNK 0 -4.001 15.677 0.000 0.00 0.00 N+0 HETATM 89 O UNK 0 -1.490 15.345 0.000 0.00 0.00 O+0 HETATM 90 C UNK 0 -7.950 8.029 0.000 0.00 0.00 C+0 HETATM 91 C UNK 0 -7.591 9.630 0.000 0.00 0.00 C+0 HETATM 92 C UNK 0 -9.258 9.149 0.000 0.00 0.00 C+0 HETATM 93 C UNK 0 -10.129 10.602 0.000 0.00 0.00 C+0 HETATM 94 N UNK 0 -9.928 12.211 0.000 0.00 0.00 N+0 HETATM 95 O UNK 0 -11.691 10.852 0.000 0.00 0.00 O+0 HETATM 96 C UNK 0 -8.712 6.691 0.000 0.00 0.00 C+0 HETATM 97 C UNK 0 -10.247 6.185 0.000 0.00 0.00 C+0 HETATM 98 C UNK 0 -11.438 5.048 0.000 0.00 0.00 C+0 HETATM 99 C UNK 0 -13.070 5.011 0.000 0.00 0.00 C+0 HETATM 100 N UNK 0 -13.968 3.731 0.000 0.00 0.00 N+0 HETATM 101 O UNK 0 -14.116 6.256 0.000 0.00 0.00 O+0 HETATM 102 C UNK 0 -6.402 0.951 0.000 0.00 0.00 C+0 HETATM 103 C UNK 0 -7.293 -0.395 0.000 0.00 0.00 C+0 HETATM 104 C UNK 0 -7.987 1.181 0.000 0.00 0.00 C+0 HETATM 105 C UNK 0 -4.992 0.333 0.000 0.00 0.00 C+0 HETATM 106 C UNK 0 -4.951 -1.279 0.000 0.00 0.00 C+0 HETATM 107 C UNK 0 -5.295 -2.899 0.000 0.00 0.00 C+0 HETATM 108 C UNK 0 -4.485 -4.324 0.000 0.00 0.00 C+0 HETATM 109 N UNK 0 -2.933 -4.528 0.000 0.00 0.00 N+0 HETATM 110 O UNK 0 -5.269 -5.740 0.000 0.00 0.00 O+0 CONECT 1 2 CONECT 2 1 3 61 76 CONECT 3 2 4 8 CONECT 4 3 5 CONECT 5 4 6 7 CONECT 6 5 CONECT 7 5 CONECT 8 3 9 10 62 CONECT 9 8 CONECT 10 8 11 CONECT 11 10 12 CONECT 12 11 13 14 CONECT 13 12 CONECT 14 12 15 CONECT 15 14 16 CONECT 16 15 17 18 CONECT 17 16 CONECT 18 16 19 CONECT 19 18 20 21 22 CONECT 20 19 CONECT 21 19 CONECT 22 19 23 CONECT 23 22 24 29 CONECT 24 23 25 27 CONECT 25 24 26 CONECT 26 25 CONECT 27 24 28 CONECT 28 27 29 31 CONECT 29 28 23 30 CONECT 30 29 CONECT 31 28 32 41 CONECT 32 31 33 CONECT 33 32 34 42 CONECT 34 33 35 41 CONECT 35 34 36 CONECT 36 35 37 38 CONECT 37 36 CONECT 38 36 39 40 CONECT 39 38 CONECT 40 38 41 CONECT 41 40 31 34 CONECT 42 33 43 61 66 CONECT 42 70 75 CONECT 43 42 44 CONECT 44 43 45 49 CONECT 45 44 46 CONECT 46 45 47 51 CONECT 47 46 48 49 CONECT 48 47 CONECT 49 47 44 50 CONECT 50 49 CONECT 51 46 52 60 CONECT 52 51 53 CONECT 53 52 54 CONECT 54 53 55 60 CONECT 55 54 56 57 CONECT 56 55 CONECT 57 55 58 CONECT 58 57 59 CONECT 59 58 60 CONECT 60 59 51 54 CONECT 61 42 2 62 CONECT 62 61 8 63 CONECT 63 62 64 65 CONECT 64 63 CONECT 65 63 66 105 CONECT 66 65 42 67 CONECT 67 66 68 102 CONECT 68 67 69 CONECT 69 68 70 96 CONECT 70 69 42 71 CONECT 71 70 72 90 CONECT 72 71 73 74 CONECT 73 72 CONECT 74 72 75 84 CONECT 75 74 42 76 CONECT 76 75 2 77 78 CONECT 77 76 CONECT 78 76 79 80 84 CONECT 79 78 CONECT 80 78 81 CONECT 81 80 82 83 CONECT 82 81 CONECT 83 81 CONECT 84 78 74 85 CONECT 85 84 86 CONECT 86 85 87 CONECT 87 86 88 89 CONECT 88 87 CONECT 89 87 CONECT 90 71 91 92 96 CONECT 91 90 CONECT 92 90 93 CONECT 93 92 94 95 CONECT 94 93 CONECT 95 93 CONECT 96 90 69 97 CONECT 97 96 98 CONECT 98 97 99 CONECT 99 98 100 101 CONECT 100 99 CONECT 101 99 CONECT 102 67 103 104 105 CONECT 103 102 CONECT 104 102 CONECT 105 102 65 106 CONECT 106 105 107 CONECT 107 106 108 CONECT 108 107 109 110 CONECT 109 108 CONECT 110 108 MASTER 0 0 0 0 0 0 0 0 110 0 248 0 END 3D PDB for HMDB0002086 (Adenosylcobalamin)COMPND HMDB0002086 HETATM 1 C1 UNL 1 6.003 11.282 0.250 1.00 0.00 C HETATM 2 C2 UNL 1 6.895 10.182 -0.155 1.00 0.00 C HETATM 3 C3 UNL 1 7.040 9.103 0.678 1.00 0.00 C HETATM 4 N1 UNL 1 7.497 7.988 0.214 1.00 0.00 N1+ HETATM 5 C4 UNL 1 7.904 7.188 1.111 1.00 0.00 C HETATM 6 C5 UNL 1 8.957 6.345 0.923 1.00 0.00 C HETATM 7 C6 UNL 1 9.566 5.954 -0.223 1.00 0.00 C HETATM 8 N2 UNL 1 9.117 6.146 -1.433 1.00 0.00 N1+ HETATM 9 C7 UNL 1 9.915 5.697 -2.353 1.00 0.00 C HETATM 10 C8 UNL 1 9.534 5.551 -3.725 1.00 0.00 C HETATM 11 C9 UNL 1 10.378 4.708 -4.590 1.00 0.00 C HETATM 12 C10 UNL 1 8.439 6.172 -4.288 1.00 0.00 C HETATM 13 N3 UNL 1 7.840 7.135 -3.634 1.00 0.00 N HETATM 14 C11 UNL 1 7.739 8.361 -4.381 1.00 0.00 C HETATM 15 C12 UNL 1 8.261 7.799 -5.772 1.00 0.00 C HETATM 16 C13 UNL 1 7.957 8.604 -7.086 1.00 0.00 C HETATM 17 C14 UNL 1 6.791 8.252 -7.929 1.00 0.00 C HETATM 18 N4 UNL 1 6.954 7.916 -9.225 1.00 0.00 N HETATM 19 O1 UNL 1 5.620 8.294 -7.493 1.00 0.00 O HETATM 20 C15 UNL 1 7.813 6.268 -5.644 1.00 0.00 C HETATM 21 C16 UNL 1 6.226 6.077 -5.566 1.00 0.00 C HETATM 22 C17 UNL 1 8.227 5.279 -6.811 1.00 0.00 C HETATM 23 C18 UNL 1 7.741 3.778 -6.685 1.00 0.00 C HETATM 24 C19 UNL 1 6.344 3.446 -7.084 1.00 0.00 C HETATM 25 O2 UNL 1 6.010 3.611 -8.281 1.00 0.00 O HETATM 26 N5 UNL 1 5.454 2.970 -6.149 1.00 0.00 N HETATM 27 C20 UNL 1 4.095 2.570 -6.374 1.00 0.00 C HETATM 28 C21 UNL 1 3.010 3.418 -5.626 1.00 0.00 C HETATM 29 C22 UNL 1 1.609 3.047 -6.213 1.00 0.00 C HETATM 30 O3 UNL 1 3.146 3.248 -4.176 1.00 0.00 O HETATM 31 P1 UNL 1 1.991 3.839 -3.220 1.00 0.00 P HETATM 32 O4 UNL 1 1.538 5.144 -3.712 1.00 0.00 O HETATM 33 O5 UNL 1 0.778 2.821 -3.268 1.00 0.00 O1- HETATM 34 O6 UNL 1 2.513 4.039 -1.715 1.00 0.00 O HETATM 35 C23 UNL 1 1.983 3.237 -0.622 1.00 0.00 C HETATM 36 C24 UNL 1 1.853 4.081 0.673 1.00 0.00 C HETATM 37 C25 UNL 1 0.876 3.505 1.739 1.00 0.00 C HETATM 38 O7 UNL 1 0.952 4.260 2.943 1.00 0.00 O HETATM 39 O8 UNL 1 3.174 4.176 1.231 1.00 0.00 O HETATM 40 C26 UNL 1 3.983 3.099 0.722 1.00 0.00 C HETATM 41 C27 UNL 1 3.045 2.230 -0.175 1.00 0.00 C HETATM 42 O9 UNL 1 3.621 1.596 -1.316 1.00 0.00 O HETATM 43 N6 UNL 1 5.139 3.606 0.020 1.00 0.00 N HETATM 44 C28 UNL 1 5.450 4.881 -0.305 1.00 0.00 C HETATM 45 N7 UNL 1 6.499 5.004 -1.153 1.00 0.00 N1+ HETATM 46 C29 UNL 1 6.839 3.703 -1.347 1.00 0.00 C HETATM 47 C30 UNL 1 7.744 3.097 -2.230 1.00 0.00 C HETATM 48 C31 UNL 1 8.083 1.688 -2.165 1.00 0.00 C HETATM 49 C32 UNL 1 9.127 1.002 -3.055 1.00 0.00 C HETATM 50 C33 UNL 1 7.388 0.897 -1.197 1.00 0.00 C HETATM 51 C34 UNL 1 7.652 -0.561 -1.025 1.00 0.00 C HETATM 52 C35 UNL 1 6.397 1.496 -0.384 1.00 0.00 C HETATM 53 C36 UNL 1 6.114 2.867 -0.515 1.00 0.00 C HETATM 54 CO1 UNL 1 7.194 7.118 -1.659 1.00 0.00 CO3- HETATM 55 C37 UNL 1 4.957 7.388 -2.328 1.00 0.00 C HETATM 56 C38 UNL 1 4.004 8.236 -1.429 1.00 0.00 C HETATM 57 O10 UNL 1 3.405 9.321 -2.170 1.00 0.00 O HETATM 58 C39 UNL 1 2.029 9.025 -2.482 1.00 0.00 C HETATM 59 N8 UNL 1 1.811 8.494 -3.812 1.00 0.00 N HETATM 60 C40 UNL 1 2.703 8.417 -4.828 1.00 0.00 C HETATM 61 N9 UNL 1 2.174 7.894 -5.949 1.00 0.00 N HETATM 62 C41 UNL 1 0.893 7.657 -5.648 1.00 0.00 C HETATM 63 C42 UNL 1 -0.154 7.125 -6.417 1.00 0.00 C HETATM 64 N10 UNL 1 0.033 6.732 -7.688 1.00 0.00 N HETATM 65 N11 UNL 1 -1.380 7.018 -5.832 1.00 0.00 N HETATM 66 C43 UNL 1 -1.605 7.410 -4.545 1.00 0.00 C HETATM 67 N12 UNL 1 -0.591 7.925 -3.796 1.00 0.00 N HETATM 68 C44 UNL 1 0.651 8.043 -4.333 1.00 0.00 C HETATM 69 C45 UNL 1 1.546 8.139 -1.307 1.00 0.00 C HETATM 70 O11 UNL 1 0.895 8.949 -0.323 1.00 0.00 O HETATM 71 C46 UNL 1 2.826 7.476 -0.767 1.00 0.00 C HETATM 72 O12 UNL 1 2.874 7.509 0.660 1.00 0.00 O HETATM 73 N13 UNL 1 7.768 9.222 -2.143 1.00 0.00 N1+ HETATM 74 C47 UNL 1 7.534 10.257 -1.406 1.00 0.00 C HETATM 75 C48 UNL 1 8.006 11.477 -2.079 1.00 0.00 C HETATM 76 C49 UNL 1 9.312 12.112 -1.471 1.00 0.00 C HETATM 77 C50 UNL 1 9.167 12.781 -0.077 1.00 0.00 C HETATM 78 C51 UNL 1 10.456 13.327 0.411 1.00 0.00 C HETATM 79 N14 UNL 1 10.635 14.634 0.679 1.00 0.00 N HETATM 80 O13 UNL 1 11.430 12.562 0.598 1.00 0.00 O HETATM 81 C52 UNL 1 7.975 11.004 -3.598 1.00 0.00 C HETATM 82 C53 UNL 1 8.966 11.804 -4.511 1.00 0.00 C HETATM 83 C54 UNL 1 6.474 11.283 -4.097 1.00 0.00 C HETATM 84 C55 UNL 1 6.112 11.245 -5.532 1.00 0.00 C HETATM 85 N15 UNL 1 6.583 12.136 -6.424 1.00 0.00 N HETATM 86 O14 UNL 1 5.253 10.438 -5.943 1.00 0.00 O HETATM 87 C56 UNL 1 8.367 9.463 -3.420 1.00 0.00 C HETATM 88 C57 UNL 1 9.918 9.191 -3.374 1.00 0.00 C HETATM 89 C58 UNL 1 11.205 5.314 -1.730 1.00 0.00 C HETATM 90 C59 UNL 1 12.428 6.261 -2.059 1.00 0.00 C HETATM 91 C60 UNL 1 13.075 6.164 -3.469 1.00 0.00 C HETATM 92 C61 UNL 1 14.408 6.803 -3.572 1.00 0.00 C HETATM 93 N16 UNL 1 14.556 8.136 -3.500 1.00 0.00 N HETATM 94 O15 UNL 1 15.437 6.112 -3.755 1.00 0.00 O HETATM 95 C62 UNL 1 10.818 5.197 -0.207 1.00 0.00 C HETATM 96 C63 UNL 1 11.888 5.761 0.804 1.00 0.00 C HETATM 97 C64 UNL 1 10.525 3.713 0.217 1.00 0.00 C HETATM 98 C65 UNL 1 7.143 7.431 2.321 1.00 0.00 C HETATM 99 C66 UNL 1 7.745 6.953 3.694 1.00 0.00 C HETATM 100 C67 UNL 1 7.545 5.438 3.993 1.00 0.00 C HETATM 101 C68 UNL 1 8.013 5.064 5.344 1.00 0.00 C HETATM 102 N17 UNL 1 9.303 5.200 5.698 1.00 0.00 N HETATM 103 O16 UNL 1 7.212 4.611 6.193 1.00 0.00 O HETATM 104 C69 UNL 1 6.808 8.963 2.146 1.00 0.00 C HETATM 105 C70 UNL 1 7.860 9.857 2.897 1.00 0.00 C HETATM 106 C71 UNL 1 5.346 9.299 2.645 1.00 0.00 C HETATM 107 C72 UNL 1 4.919 8.741 3.951 1.00 0.00 C HETATM 108 N18 UNL 1 4.995 9.461 5.087 1.00 0.00 N HETATM 109 O17 UNL 1 4.446 7.583 4.029 1.00 0.00 O HETATM 110 H1 UNL 1 6.387 11.827 1.109 1.00 0.00 H HETATM 111 H2 UNL 1 5.795 12.013 -0.523 1.00 0.00 H HETATM 112 H3 UNL 1 5.018 10.887 0.485 1.00 0.00 H HETATM 113 H4 UNL 1 9.364 6.021 1.759 1.00 0.00 H HETATM 114 H5 UNL 1 11.167 4.163 -4.098 1.00 0.00 H HETATM 115 H6 UNL 1 10.858 5.316 -5.356 1.00 0.00 H HETATM 116 H7 UNL 1 9.826 3.915 -5.057 1.00 0.00 H HETATM 117 H8 UNL 1 6.705 8.649 -4.514 1.00 0.00 H HETATM 118 H9 UNL 1 9.363 7.856 -5.746 1.00 0.00 H HETATM 119 H10 UNL 1 8.843 8.531 -7.729 1.00 0.00 H HETATM 120 H11 UNL 1 7.888 9.659 -6.838 1.00 0.00 H HETATM 121 H12 UNL 1 7.845 7.887 -9.612 1.00 0.00 H HETATM 122 H13 UNL 1 6.182 7.710 -9.775 1.00 0.00 H HETATM 123 H14 UNL 1 5.976 5.238 -4.914 1.00 0.00 H HETATM 124 H15 UNL 1 5.768 5.917 -6.543 1.00 0.00 H HETATM 125 H16 UNL 1 5.680 6.920 -5.178 1.00 0.00 H HETATM 126 H17 UNL 1 9.304 5.300 -6.951 1.00 0.00 H HETATM 127 H18 UNL 1 7.842 5.669 -7.754 1.00 0.00 H HETATM 128 H19 UNL 1 8.405 3.153 -7.288 1.00 0.00 H HETATM 129 H20 UNL 1 7.842 3.455 -5.651 1.00 0.00 H HETATM 130 H21 UNL 1 5.790 2.864 -5.245 1.00 0.00 H HETATM 131 H22 UNL 1 3.889 2.563 -7.448 1.00 0.00 H HETATM 132 H23 UNL 1 4.015 1.536 -6.037 1.00 0.00 H HETATM 133 H24 UNL 1 3.195 4.472 -5.864 1.00 0.00 H HETATM 134 H25 UNL 1 1.324 2.034 -5.918 1.00 0.00 H HETATM 135 H26 UNL 1 1.609 3.109 -7.303 1.00 0.00 H HETATM 136 H27 UNL 1 0.845 3.740 -5.862 1.00 0.00 H HETATM 137 H28 UNL 1 1.031 2.744 -0.841 1.00 0.00 H HETATM 138 H29 UNL 1 1.500 5.085 0.426 1.00 0.00 H HETATM 139 H30 UNL 1 -0.150 3.544 1.366 1.00 0.00 H HETATM 140 H31 UNL 1 1.117 2.470 1.987 1.00 0.00 H HETATM 141 H32 UNL 1 0.404 3.756 3.581 1.00 0.00 H HETATM 142 H33 UNL 1 4.323 2.511 1.579 1.00 0.00 H HETATM 143 H34 UNL 1 2.585 1.442 0.428 1.00 0.00 H HETATM 144 H35 UNL 1 4.196 0.889 -0.961 1.00 0.00 H HETATM 145 H36 UNL 1 4.939 5.685 0.042 1.00 0.00 H HETATM 146 H37 UNL 1 8.168 3.648 -2.950 1.00 0.00 H HETATM 147 H38 UNL 1 9.911 0.555 -2.442 1.00 0.00 H HETATM 148 H39 UNL 1 8.658 0.232 -3.669 1.00 0.00 H HETATM 149 H40 UNL 1 9.594 1.729 -3.719 1.00 0.00 H HETATM 150 H41 UNL 1 7.473 -1.082 -1.967 1.00 0.00 H HETATM 151 H42 UNL 1 7.013 -1.024 -0.270 1.00 0.00 H HETATM 152 H43 UNL 1 8.690 -0.718 -0.722 1.00 0.00 H HETATM 153 H44 UNL 1 5.872 0.941 0.295 1.00 0.00 H HETATM 154 H45 UNL 1 5.024 7.906 -3.266 1.00 0.00 H HETATM 155 H46 UNL 1 4.537 6.408 -2.557 1.00 0.00 H HETATM 156 H47 UNL 1 4.618 8.684 -0.657 1.00 0.00 H HETATM 157 H48 UNL 1 1.488 9.977 -2.462 1.00 0.00 H HETATM 158 H49 UNL 1 3.665 8.736 -4.786 1.00 0.00 H HETATM 159 H50 UNL 1 0.906 6.820 -8.101 1.00 0.00 H HETATM 160 H51 UNL 1 -0.705 6.359 -8.195 1.00 0.00 H HETATM 161 H52 UNL 1 -2.536 7.313 -4.142 1.00 0.00 H HETATM 162 H53 UNL 1 0.831 7.377 -1.626 1.00 0.00 H HETATM 163 H54 UNL 1 0.564 8.319 0.351 1.00 0.00 H HETATM 164 H55 UNL 1 2.829 6.432 -1.064 1.00 0.00 H HETATM 165 H56 UNL 1 3.701 7.048 0.906 1.00 0.00 H HETATM 166 H57 UNL 1 7.306 12.302 -1.971 1.00 0.00 H HETATM 167 H58 UNL 1 10.098 11.376 -1.378 1.00 0.00 H HETATM 168 H59 UNL 1 9.668 12.896 -2.144 1.00 0.00 H HETATM 169 H60 UNL 1 8.410 13.571 -0.132 1.00 0.00 H HETATM 170 H61 UNL 1 8.833 12.040 0.647 1.00 0.00 H HETATM 171 H62 UNL 1 11.503 14.947 0.987 1.00 0.00 H HETATM 172 H63 UNL 1 9.906 15.264 0.565 1.00 0.00 H HETATM 173 H64 UNL 1 8.668 12.852 -4.577 1.00 0.00 H HETATM 174 H65 UNL 1 8.994 11.377 -5.512 1.00 0.00 H HETATM 175 H66 UNL 1 9.988 11.783 -4.133 1.00 0.00 H HETATM 176 H67 UNL 1 5.800 10.609 -3.559 1.00 0.00 H HETATM 177 H68 UNL 1 6.163 12.292 -3.807 1.00 0.00 H HETATM 178 H69 UNL 1 7.195 12.834 -6.141 1.00 0.00 H HETATM 179 H70 UNL 1 6.306 12.085 -7.354 1.00 0.00 H HETATM 180 H71 UNL 1 10.421 9.696 -2.568 1.00 0.00 H HETATM 181 H72 UNL 1 10.151 8.149 -3.225 1.00 0.00 H HETATM 182 H73 UNL 1 10.393 9.480 -4.311 1.00 0.00 H HETATM 183 H74 UNL 1 11.507 4.311 -2.025 1.00 0.00 H HETATM 184 H75 UNL 1 13.242 5.993 -1.388 1.00 0.00 H HETATM 185 H76 UNL 1 12.159 7.299 -1.849 1.00 0.00 H HETATM 186 H77 UNL 1 13.200 5.110 -3.725 1.00 0.00 H HETATM 187 H78 UNL 1 12.426 6.632 -4.206 1.00 0.00 H HETATM 188 H79 UNL 1 15.438 8.530 -3.600 1.00 0.00 H HETATM 189 H80 UNL 1 13.787 8.707 -3.358 1.00 0.00 H HETATM 190 H81 UNL 1 11.580 5.639 1.843 1.00 0.00 H HETATM 191 H82 UNL 1 12.832 5.223 0.704 1.00 0.00 H HETATM 192 H83 UNL 1 12.068 6.825 0.639 1.00 0.00 H HETATM 193 H84 UNL 1 10.155 3.653 1.244 1.00 0.00 H HETATM 194 H85 UNL 1 11.434 3.111 0.148 1.00 0.00 H HETATM 195 H86 UNL 1 9.780 3.256 -0.430 1.00 0.00 H HETATM 196 H87 UNL 1 6.211 6.868 2.179 1.00 0.00 H HETATM 197 H88 UNL 1 8.808 7.205 3.739 1.00 0.00 H HETATM 198 H89 UNL 1 7.260 7.495 4.506 1.00 0.00 H HETATM 199 H90 UNL 1 6.482 5.198 3.902 1.00 0.00 H HETATM 200 H91 UNL 1 8.077 4.825 3.264 1.00 0.00 H HETATM 201 H92 UNL 1 9.590 4.955 6.591 1.00 0.00 H HETATM 202 H93 UNL 1 9.949 5.548 5.064 1.00 0.00 H HETATM 203 H94 UNL 1 7.707 10.916 2.702 1.00 0.00 H HETATM 204 H95 UNL 1 8.879 9.606 2.588 1.00 0.00 H HETATM 205 H96 UNL 1 7.795 9.727 3.978 1.00 0.00 H HETATM 206 H97 UNL 1 5.223 10.381 2.724 1.00 0.00 H HETATM 207 H98 UNL 1 4.625 8.958 1.903 1.00 0.00 H HETATM 208 H99 UNL 1 4.695 9.076 5.926 1.00 0.00 H HETATM 209 HA0 UNL 1 5.345 10.366 5.069 1.00 0.00 H CONECT 1 2 110 111 112 CONECT 2 3 3 74 CONECT 3 4 104 CONECT 4 5 5 54 CONECT 5 6 98 CONECT 6 7 7 113 CONECT 7 8 95 CONECT 8 9 9 54 CONECT 9 10 89 CONECT 10 11 12 12 CONECT 11 114 115 116 CONECT 12 13 20 CONECT 13 14 54 CONECT 14 15 87 117 CONECT 15 16 20 118 CONECT 16 17 119 120 CONECT 17 18 19 19 CONECT 18 121 122 CONECT 20 21 22 CONECT 21 123 124 125 CONECT 22 23 126 127 CONECT 23 24 128 129 CONECT 24 25 25 26 CONECT 26 27 130 CONECT 27 28 131 132 CONECT 28 29 30 133 CONECT 29 134 135 136 CONECT 30 31 CONECT 31 32 32 33 34 CONECT 34 35 CONECT 35 36 41 137 CONECT 36 37 39 138 CONECT 37 38 139 140 CONECT 38 141 CONECT 39 40 CONECT 40 41 43 142 CONECT 41 42 143 CONECT 42 144 CONECT 43 44 53 CONECT 44 45 45 145 CONECT 45 46 54 CONECT 46 47 47 53 CONECT 47 48 146 CONECT 48 49 50 50 CONECT 49 147 148 149 CONECT 50 51 52 CONECT 51 150 151 152 CONECT 52 53 53 153 CONECT 54 55 73 CONECT 55 56 154 155 CONECT 56 57 71 156 CONECT 57 58 CONECT 58 59 69 157 CONECT 59 60 68 CONECT 60 61 61 158 CONECT 61 62 CONECT 62 63 63 68 CONECT 63 64 65 CONECT 64 159 160 CONECT 65 66 66 CONECT 66 67 161 CONECT 67 68 68 CONECT 69 70 71 162 CONECT 70 163 CONECT 71 72 164 CONECT 72 165 CONECT 73 74 74 87 CONECT 74 75 CONECT 75 76 81 166 CONECT 76 77 167 168 CONECT 77 78 169 170 CONECT 78 79 80 80 CONECT 79 171 172 CONECT 81 82 83 87 CONECT 82 173 174 175 CONECT 83 84 176 177 CONECT 84 85 86 86 CONECT 85 178 179 CONECT 87 88 CONECT 88 180 181 182 CONECT 89 90 95 183 CONECT 90 91 184 185 CONECT 91 92 186 187 CONECT 92 93 94 94 CONECT 93 188 189 CONECT 95 96 97 CONECT 96 190 191 192 CONECT 97 193 194 195 CONECT 98 99 104 196 CONECT 99 100 197 198 CONECT 100 101 199 200 CONECT 101 102 103 103 CONECT 102 201 202 CONECT 104 105 106 CONECT 105 203 204 205 CONECT 106 107 206 207 CONECT 107 108 109 109 CONECT 108 208 209 END SMILES for HMDB0002086 (Adenosylcobalamin)[C@H]1(C[Co-3]2345[N+]6=C7C(C)=C8N2[C@]([H])([C@H](CC(=O)N)[C@]8(CCC(=O)NC[C@@H](C)OP([O-])(=O)O[C@@H]2[C@@H](CO)O[C@H](N8C=[N+]3C3=CC(C)=C(C)C=C83)[C@@H]2O)C)[C@@]2(C)[C@@](CC(=O)N)([C@H](CCC(=O)N)C(C(C)=C3[N+]4=C(C=C6C([C@@H]7CCC(=O)N)(C)C)[C@@H](CCC(=O)N)[C@@]3(CC(=O)N)C)=[N+]52)C)O[C@@H](N2C3=NC=NC(=C3N=C2)N)[C@H](O)[C@@H]1O INCHI for HMDB0002086 (Adenosylcobalamin)InChI=1S/C62H90N13O14P.C10H12N5O3.Co/c1-29-20-39-40(21-30(29)2)75(28-70-39)57-52(84)53(41(27-76)87-57)89-90(85,86)88-31(3)26-69-49(83)18-19-59(8)37(22-46(66)80)56-62(11)61(10,25-48(68)82)36(14-17-45(65)79)51(74-62)33(5)55-60(9,24-47(67)81)34(12-15-43(63)77)38(71-55)23-42-58(6,7)35(13-16-44(64)78)50(72-42)32(4)54(59)73-56;1-4-6(16)7(17)10(18-4)15-3-14-5-8(11)12-2-13-9(5)15;/h20-21,23,28,31,34-37,41,52-53,56-57,76,84H,12-19,22,24-27H2,1-11H3,(H15,63,64,65,66,67,68,69,71,72,73,74,77,78,79,80,81,82,83,85,86);2-4,6-7,10,16-17H,1H2,(H2,11,12,13);/q;;+2/p-2/t31-,34-,35-,36-,37+,41-,52-,53-,56-,57+,59-,60+,61+,62+;4-,6-,7-,10-;/m11./s1 3D Structure for HMDB0002086 (Adenosylcobalamin) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C72H100CoN18O17P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 1579.5818 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 1578.65834557 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | (10S,12R,13S,17R,23R,24R,25R,30S,35S,36S,40S,41S,42R,46R)-1-{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}-30,35,40-tris(2-carbamoylethyl)-24,36,41-tris(carbamoylmethyl)-46-hydroxy-12-(hydroxymethyl)-5,6,17,23,28,31,31,36,38,41,42-undecamethyl-15,20-dioxo-11,14,16-trioxa-2lambda5,9,19,26,43lambda5,44lambda5,45lambda5-heptaaza-15lambda5-phospha-1-cobaltadodecacyclo[27.14.1.1^{1,34}.1^{2,9}.1^{10,13}.0^{1,26}.0^{3,8}.0^{23,27}.0^{25,42}.0^{32,44}.0^{39,43}.0^{37,45}]heptatetraconta-2(47),3,5,7,27,29(44),32,34(45),37,39(43)-decaene-2,43,44,45-tetrakis(ylium)-1,1,1-triuid-15-olate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | (10S,12R,13S,17R,23R,24R,25R,30S,35S,36S,40S,41S,42R,46R)-1-{[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}-30,35,40-tris(2-carbamoylethyl)-24,36,41-tris(carbamoylmethyl)-46-hydroxy-12-(hydroxymethyl)-5,6,17,23,28,31,31,36,38,41,42-undecamethyl-15,20-dioxo-11,14,16-trioxa-2lambda5,9,19,26,43lambda5,44lambda5,45lambda5-heptaaza-15lambda5-phospha-1-cobaltadodecacyclo[27.14.1.1^{1,34}.1^{2,9}.1^{10,13}.0^{1,26}.0^{3,8}.0^{23,27}.0^{25,42}.0^{32,44}.0^{39,43}.0^{37,45}]heptatetraconta-2(47),3,5,7,27,29(44),32,34(45),37,39(43)-decaene-2,43,44,45-tetrakis(ylium)-1,1,1-triuid-15-olate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 13870-90-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [C@H]1(C[Co-3]2345[N+]6=C7C(C)=C8N2[C@]([H])([C@H](CC(=O)N)[C@]8(CCC(=O)NC[C@@H](C)OP([O-])(=O)O[C@@H]2[C@@H](CO)O[C@H](N8C=[N+]3C3=CC(C)=C(C)C=C83)[C@@H]2O)C)[C@@]2(C)[C@@](CC(=O)N)([C@H](CCC(=O)N)C(C(C)=C3[N+]4=C(C=C6C([C@@H]7CCC(=O)N)(C)C)[C@@H](CCC(=O)N)[C@@]3(CC(=O)N)C)=[N+]52)C)O[C@@H](N2C3=NC=NC(=C3N=C2)N)[C@H](O)[C@@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C62H90N13O14P.C10H12N5O3.Co/c1-29-20-39-40(21-30(29)2)75(28-70-39)57-52(84)53(41(27-76)87-57)89-90(85,86)88-31(3)26-69-49(83)18-19-59(8)37(22-46(66)80)56-62(11)61(10,25-48(68)82)36(14-17-45(65)79)51(74-62)33(5)55-60(9,24-47(67)81)34(12-15-43(63)77)38(71-55)23-42-58(6,7)35(13-16-44(64)78)50(72-42)32(4)54(59)73-56;1-4-6(16)7(17)10(18-4)15-3-14-5-8(11)12-2-13-9(5)15;/h20-21,23,28,31,34-37,41,52-53,56-57,76,84H,12-19,22,24-27H2,1-11H3,(H15,63,64,65,66,67,68,69,71,72,73,74,77,78,79,80,81,82,83,85,86);2-4,6-7,10,16-17H,1H2,(H2,11,12,13);/q;;+2/p-2/t31-,34-,35-,36-,37+,41-,52-,53-,56-,57+,59-,60+,61+,62+;4-,6-,7-,10-;/m11./s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | ZIHHMGTYZOSFRC-OUCXYWSSSA-L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Belongs to the class of organic compounds known as cobalamin derivatives. These are organic compounds containing a corrin ring, a cobalt atom, an a nucleotide moiety. Cobalamin Derivatives are actually derived from vitamin B12. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Tetrapyrroles and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Corrinoids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Cobalamin derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physiological effect | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disposition | Biological location
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Process | Naturally occurring process
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Role | Biological role
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Molecular Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Chromatographic Properties | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Molecular Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Chromatographic Properties | Predicted Retention TimesUnderivatized
Predicted Kovats Retention IndicesNot Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Associated Disorders and Diseases | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disease References | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Associated OMIM IDs | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB022837 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | ADENOSYLCOBALAMIN | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Cobamamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 18408 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Food Biomarker Ontology | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| VMH ID | ADOCBL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MarkerDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Good Scents ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in isomerase activity
- Specific function:
- Involved in the degradation of several amino acids, odd-chain fatty acids and cholesterol via propionyl-CoA to the tricarboxylic acid cycle. MCM has different functions in other species.
- Gene Name:
- MUT
- Uniprot ID:
- P22033
- Molecular weight:
- 83133.755
- General function:
- Involved in ATP binding
- Specific function:
- Not Available
- Gene Name:
- MMAB
- Uniprot ID:
- Q96EY8
- Molecular weight:
- 27387.975
- General function:
- Involved in nucleotide binding
- Specific function:
- Probable GTPase. May function as chaperone. May be involved in the transport of cobalamin (Cbl) into mitochondria for the final steps of adenosylcobalamin (AdoCbl) synthesis
- Gene Name:
- MMAA
- Uniprot ID:
- Q8IVH4
- Molecular weight:
- 46537.9