| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:48:59 UTC |
|---|
| HMDB ID | HMDB0000086 |
|---|
| Secondary Accession Numbers | - HMDB0000049
- HMDB00049
- HMDB00086
|
|---|
| Metabolite Identification |
|---|
| Common Name | Glycerophosphocholine |
|---|
| Description | Glycerophosphorylcholine (GPC) is a choline derivative and one of the two major forms of choline storage (along with phosphocholine) in the cytosol. Glycerophosphorylcholine is also one of the four major organic osmolytes in renal medullary cells, changing their intracellular osmolyte concentration in parallel with extracellular tonicity during cellular osmoadaptation. As an osmolyte, Glycerophosphorylcholine counteracts the effects of urea on enzymes and other macromolecules. Kidneys (especially medullar cells), which are exposed under normal physiological conditions to widely fluctuating extracellular solute concentrations, respond to hypertonic stress by accumulating the organic osmolytes glycerophosphorylcholine (GPC), betaine, myo-inositol, sorbitol and free amino acids. Increased intracellular contents of these osmolytes are achieved by a combination of increased uptake (myo-inositol and betaine) and synthesis (sorbitol, GPC), decreased degradation (GPC) and reduced osmolyte release. GPC is formed in the breakdown of phosphatidylcholine (PtC). This pathway is active in many body tissues, including mammary tissue. |
|---|
| Structure | C[N+](C)(C)CCOP([O-])(=O)OC[C@H](O)CO InChI=1S/C8H20NO6P/c1-9(2,3)4-5-14-16(12,13)15-7-8(11)6-10/h8,10-11H,4-7H2,1-3H3/t8-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| (2R)-2,3-Dihydroxypropyl 2-(trimethylammonio)ethyl phosphate | ChEBI | | Alfoscerate de choline | ChEBI | | Alfoscerato de colina | ChEBI | | alpha-Glycerophosphorylcholine | ChEBI | | Choline alphoscerate | ChEBI | | Choline glycerophosphate | ChEBI | | Cholini alfosceras | ChEBI | | Cholini glycerophosphas | ChEBI | | Glicerofosfato de colina | ChEBI | | Glycerol phosphorylcholine | ChEBI | | Glycerol-3-phosphatidylcholine | ChEBI | | Glycerol-3-phosphocholine | ChEBI | | Glycerophosphate de choline | ChEBI | | Glycerophosphorylcholine | ChEBI | | GPCho | ChEBI | | L-alpha-Glycerophosphocholine | ChEBI | | L-alpha-Glycerophosphorylcholine | ChEBI | | L-Choline hydroxide 2,3-dihydroxypropyl hydrogen phosphate inner salt | ChEBI | | sn-3-GPC | ChEBI | | sn-Glycero-3-phosphocholine | ChEBI | | sn-Glycerol 3-phosphocholine | ChEBI | | Gliatilin | Kegg | | (2R)-2,3-Dihydroxypropyl 2-(trimethylammonio)ethyl phosphoric acid | Generator | | Alfosceric acid de choline | Generator | | a-Glycerophosphorylcholine | Generator | | Α-glycerophosphorylcholine | Generator | | Choline alphosceric acid | Generator | | Choline glycerophosphoric acid | Generator | | Glycerophosphoric acid de choline | Generator | | L-a-Glycerophosphocholine | Generator | | L-Α-glycerophosphocholine | Generator | | L-a-Glycerophosphorylcholine | Generator | | L-Α-glycerophosphorylcholine | Generator | | L-Choline hydroxide 2,3-dihydroxypropyl hydrogen phosphoric acid inner salt | Generator | | Choline alfosceric acid | HMDB | | Glycerol 3 phosphocholine | HMDB | | Glycerylphosphorylcholine | HMDB | | Alphoscerate, choline | HMDB | | L alpha Glycerylphosphorylcholine | HMDB | | 3-Phosphocholine, glycerol | HMDB | | Glycerol 3-phosphocholine | HMDB | | L-alpha-Glycerylphosphorylcholine | HMDB | | Alfoscerate, choline | HMDB | | Glycerophosphate, choline | HMDB | | 2-[[(2,3-Dihydroxypropoxy)hydroxyphosphinyl]oxy]-N,N,N-trimethyl-ethanaminium inner salt | HMDB | | a-Glycerylphosphorylcholine | HMDB | | alpha-Glycerylphosphorylcholine | HMDB | | Choline alfoscerate | HMDB | | Glycerophosphatidylcholine | HMDB | | GPC | HMDB | | Hydrogen glycerophosphate choline | HMDB | | Cereton | HMDB | | Cholicerin | HMDB | | Cholitiline | HMDB | | Delecit | HMDB | | Glycerol 3-phosphorylcholine | HMDB | | Glycerophosphoric acid choline ester | HMDB | | Glyceryl 3-phosphorylcholine | HMDB | | Glycerylphosphocholine | HMDB | | L-alpha-GPC | HMDB | | L-Α-GPC | HMDB | | L-Α-glycerylphosphorylcholine | HMDB | | O-(sn-Glycero-3-phosphoryl)-choline | HMDB | | sn-Glycero-3-phosphorylcholine | HMDB | | Α-glycerylphosphorylcholine | HMDB | | Glycerophosphocholine | ChEBI |
| Show more...
|---|
| Chemical Formula | C8H20NO6P |
|---|
| Average Molecular Weight | 257.223 |
|---|
| Monoisotopic Molecular Weight | 257.102824366 |
|---|
| IUPAC Name | (2-{[(2R)-2,3-dihydroxypropyl phosphono]oxy}ethyl)trimethylazanium |
|---|
| Traditional Name | (2-{[(2R)-2,3-dihydroxypropyl phosphono]oxy}ethyl)trimethylazanium |
|---|
| CAS Registry Number | 28319-77-9 |
|---|
| SMILES | C[N+](C)(C)CCOP([O-])(=O)OC[C@H](O)CO |
|---|
| InChI Identifier | InChI=1S/C8H20NO6P/c1-9(2,3)4-5-14-16(12,13)15-7-8(11)6-10/h8,10-11H,4-7H2,1-3H3/t8-/m1/s1 |
|---|
| InChI Key | SUHOQUVVVLNYQR-MRVPVSSYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as glycerophosphocholines. These are lipids containing a glycerol moiety carrying a phosphocholine at the 3-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerophospholipids |
|---|
| Sub Class | Glycerophosphocholines |
|---|
| Direct Parent | Glycerophosphocholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycero-3-phosphocholine
- Phosphocholine
- Dialkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Tetraalkylammonium salt
- Quaternary ammonium salt
- 1,2-diol
- Secondary alcohol
- Organic nitrogen compound
- Organic salt
- Hydrocarbon derivative
- Primary alcohol
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Amine
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 142.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | | Show more...
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Glycerophosphocholine,1TMS,isomer #1 | C[N+](C)(C)CCOP(=O)([O-])OC[C@@H](CO)O[Si](C)(C)C | 1806.4 | Semi standard non polar | 33892256 | | Glycerophosphocholine,1TMS,isomer #2 | C[N+](C)(C)CCOP(=O)([O-])OC[C@H](O)CO[Si](C)(C)C | 1808.9 | Semi standard non polar | 33892256 | | Glycerophosphocholine,2TMS,isomer #1 | C[N+](C)(C)CCOP(=O)([O-])OC[C@@H](CO[Si](C)(C)C)O[Si](C)(C)C | 1818.7 | Semi standard non polar | 33892256 | | Glycerophosphocholine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)O[C@H](CO)COP(=O)([O-])OCC[N+](C)(C)C | 2035.3 | Semi standard non polar | 33892256 | | Glycerophosphocholine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC[C@@H](O)COP(=O)([O-])OCC[N+](C)(C)C | 2042.6 | Semi standard non polar | 33892256 | | Glycerophosphocholine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC[C@H](COP(=O)([O-])OCC[N+](C)(C)C)O[Si](C)(C)C(C)(C)C | 2318.1 | Semi standard non polar | 33892256 |
| Show more...
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (Non-derivatized) - 70eV, Positive | splash10-001j-7910000000-79a0f6ec434740d09410 | 2017-08-28 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (TMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Glycerophosphocholine GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 20V, Negative-QTOF | splash10-004i-9310000000-e2633e56c86952054710 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 10V, Negative-QTOF | splash10-0a4i-1690000000-53ffc11b39ba8f89f803 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 40V, Negative-QTOF | splash10-004i-9000000000-d4d2c5d7e0856e948624 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 20V, Negative-QTOF | splash10-004i-9400000000-848201d8ac3bd454c67a | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 40V, Positive-QTOF | splash10-01w1-9300000000-987e5431e2d611fa5032 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 10V, Negative-QTOF | splash10-0a4i-2490000000-efb55f1ddbed9f1a8754 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 20V, Positive-QTOF | splash10-0udi-1900000000-df7b0a6ce3c788063f53 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Glycerophosphocholine 10V, Positive-QTOF | splash10-0zfr-0960000000-7df186ffc569be161b39 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 10V, Positive-QTOF | splash10-052r-9140000000-3347b58fbc5fe0b0a201 | 2019-02-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 20V, Positive-QTOF | splash10-0a7r-9210000000-60118aed826a14bb4cb1 | 2019-02-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 40V, Positive-QTOF | splash10-0a4r-9000000000-b6ac875e42841f04bcfc | 2019-02-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 10V, Negative-QTOF | splash10-0a4i-0390000000-9adc870a20cd016e3aa2 | 2019-02-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 20V, Negative-QTOF | splash10-0kor-2920000000-b31eb4ca99ba45d9f931 | 2019-02-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 40V, Negative-QTOF | splash10-004i-9100000000-dec05a487d7ae73f1d27 | 2019-02-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 10V, Negative-QTOF | splash10-0a4i-0090000000-bd45c4f9a61bbdbbd2d6 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 20V, Negative-QTOF | splash10-0a4i-3960000000-2d618fc6f1a9c434909c | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 40V, Negative-QTOF | splash10-004i-9000000000-a5e502a2627af2048a1f | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 10V, Positive-QTOF | splash10-001i-0960000000-782cafcc84f2e1b58561 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 20V, Positive-QTOF | splash10-001i-0900000000-5fbae562ef71fe1bd552 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Glycerophosphocholine 40V, Positive-QTOF | splash10-0079-9800000000-00d2deed17b9ac0f5789 | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum |
| Show more...
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biospecimen Locations | - Blood
- Breast Milk
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Semen
- Urine
|
|---|
| Tissue Locations | - Brain
- Kidney
- Placenta
- Prostate
- Spleen
|
|---|
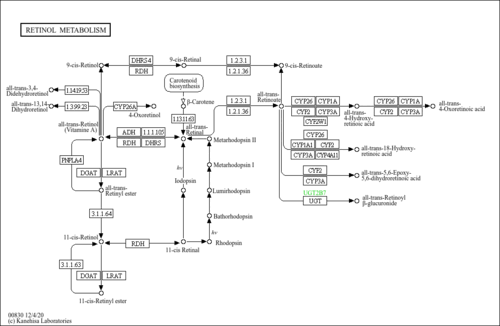
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 34.0 +/- 5.0 uM | Infant (0-1 year old) | Both | Normal | | details | | Blood | Detected and Quantified | 40.0 +/- 4.0 uM | Newborn (0-30 days old) | Both | Normal | | details | | Blood | Detected and Quantified | 29.0 +/- 5.0 uM | Infant (0-1 year old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 33.0 +/- 4.0 uM | Adult (>18 years old) | Female | Normal | | details | | Breast Milk | Detected and Quantified | 471 +/- 161 uM | Adult (>18 years old) | Female | Normal | | details | | Breast Milk | Detected and Quantified | 362 +/- 70 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 3.94 +/- 1.17 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 3.9 +/- 1.2 uM | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 1.05 +/- 0.248 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 1.38 +/- 0.531 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected and Quantified | 1.84 +/- 0.652 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 1.98 +/- 1.45 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected and Quantified | 2.53 +/- 4.35 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 9.82 +/- 6.76 uM | Adult (>18 years old) | Both | Normal | | details | | Semen | Detected and Quantified | 2.5 +/- 1.6 uM | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details |
| Show more...
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Kidney cancer | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 4.03 +/- 1.52 uM | Elderly (>65 years old) | Not Specified | Vascular dementia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 4.03 (2.51-5.55) uM | Adult (>18 years old) | Both | Vascular dementia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 6.9 +/- 3.2 uM | Elderly (>65 years old) | Both | Alzheimer's disease | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Attachment loss | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Missing teeth | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Periodontal Probing Depth | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Prosthesis/Missing teeth | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Oral cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Breast cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Pancreatic cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Periodontal diseases | | details |
| Show more...
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Kidney cancer |
|---|
- Lin L, Huang Z, Gao Y, Chen Y, Hang W, Xing J, Yan X: LC-MS-based serum metabolic profiling for genitourinary cancer classification and cancer type-specific biomarker discovery. Proteomics. 2012 Aug;12(14):2238-46. doi: 10.1002/pmic.201200016. [PubMed:22685041 ]
| | Multi-infarct dementia |
|---|
- Walter A, Korth U, Hilgert M, Hartmann J, Weichel O, Hilgert M, Fassbender K, Schmitt A, Klein J: Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol Aging. 2004 Nov-Dec;25(10):1299-303. [PubMed:15465626 ]
| | Alzheimer's disease |
|---|
- Walter A, Korth U, Hilgert M, Hartmann J, Weichel O, Hilgert M, Fassbender K, Schmitt A, Klein J: Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol Aging. 2004 Nov-Dec;25(10):1299-303. [PubMed:15465626 ]
| | Perillyl alcohol administration for cancer treatment |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Pancreatic cancer |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Periodontal disease |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Attachment loss |
|---|
- Liebsch C, Pitchika V, Pink C, Samietz S, Kastenmuller G, Artati A, Suhre K, Adamski J, Nauck M, Volzke H, Friedrich N, Kocher T, Holtfreter B, Pietzner M: The Saliva Metabolome in Association to Oral Health Status. J Dent Res. 2019 Jun;98(6):642-651. doi: 10.1177/0022034519842853. Epub 2019 Apr 26. [PubMed:31026179 ]
| | Missing teeth |
|---|
- Liebsch C, Pitchika V, Pink C, Samietz S, Kastenmuller G, Artati A, Suhre K, Adamski J, Nauck M, Volzke H, Friedrich N, Kocher T, Holtfreter B, Pietzner M: The Saliva Metabolome in Association to Oral Health Status. J Dent Res. 2019 Jun;98(6):642-651. doi: 10.1177/0022034519842853. Epub 2019 Apr 26. [PubMed:31026179 ]
| | Periodontal Probing Depth |
|---|
- Liebsch C, Pitchika V, Pink C, Samietz S, Kastenmuller G, Artati A, Suhre K, Adamski J, Nauck M, Volzke H, Friedrich N, Kocher T, Holtfreter B, Pietzner M: The Saliva Metabolome in Association to Oral Health Status. J Dent Res. 2019 Jun;98(6):642-651. doi: 10.1177/0022034519842853. Epub 2019 Apr 26. [PubMed:31026179 ]
| | Prosthesis/Missing teeth |
|---|
- Liebsch C, Pitchika V, Pink C, Samietz S, Kastenmuller G, Artati A, Suhre K, Adamski J, Nauck M, Volzke H, Friedrich N, Kocher T, Holtfreter B, Pietzner M: The Saliva Metabolome in Association to Oral Health Status. J Dent Res. 2019 Jun;98(6):642-651. doi: 10.1177/0022034519842853. Epub 2019 Apr 26. [PubMed:31026179 ]
|
|
|---|
| Associated OMIM IDs | |
|---|
| External Links |
|---|
| DrugBank ID | DB04660 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB030952 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 571409 |
|---|
| KEGG Compound ID | C00670 |
|---|
| BioCyc ID | L-1-GLYCERO-PHOSPHORYLCHOLINE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Alpha-GPC |
|---|
| METLIN ID | 370 |
|---|
| PubChem Compound | 657272 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16870 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | G3PC |
|---|
| MarkerDB ID | MDB00000045 |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Evans, Christopher Thomas; McCague, Raymond; Tyrrell, Nicholas David. Preparation of phospholipid-intermediate glycerophosphocholine by a crystallization process. PCT Int. Appl. (1993), 8 pp. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| General References | - Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Satlin A, Bodick N, Offen WW, Renshaw PF: Brain proton magnetic resonance spectroscopy (1H-MRS) in Alzheimer's disease: changes after treatment with xanomeline, an M1 selective cholinergic agonist. Am J Psychiatry. 1997 Oct;154(10):1459-61. [PubMed:9326834 ]
- Walter A, Korth U, Hilgert M, Hartmann J, Weichel O, Hilgert M, Fassbender K, Schmitt A, Klein J: Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol Aging. 2004 Nov-Dec;25(10):1299-303. [PubMed:15465626 ]
- Theberge J, Al-Semaan Y, Jensen JE, Williamson PC, Neufeld RW, Menon RS, Schaefer B, Densmore M, Drost DJ: Comparative study of proton and phosphorus magnetic resonance spectroscopy in schizophrenia at 4 Tesla. Psychiatry Res. 2004 Nov 15;132(1):33-9. [PubMed:15546701 ]
- Miller BL, Lin KM, Djenderedjian A, Tang C, Hill E, Fu P, Nuccio C, Jenden DJ: Changes in red blood cell choline and choline-bound lipids with oral lithium. Experientia. 1990 May 15;46(5):454-6. [PubMed:2347393 ]
- Caetano SC, Fonseca M, Olvera RL, Nicoletti M, Hatch JP, Stanley JA, Hunter K, Lafer B, Pliszka SR, Soares JC: Proton spectroscopy study of the left dorsolateral prefrontal cortex in pediatric depressed patients. Neurosci Lett. 2005 Aug 26;384(3):321-6. [PubMed:15936878 ]
- Nitsch R, Pittas A, Blusztajn JK, Slack BE, Growdon JH, Wurtman RJ: Alterations of phospholipid metabolites in postmortem brain from patients with Alzheimer's disease. Ann N Y Acad Sci. 1991;640:110-3. [PubMed:1663712 ]
- Chap HJ, Moatti JP, Mieusset R, Nieto M, Laneelle G, Bennet PJ, Mansat A, Pontonnier F, Douste-Blazy L: Simple, rapid enzymatic determination of glycerophosphocholine in human seminal plasma. Clin Chem. 1988 Jan;34(1):106-9. [PubMed:2827914 ]
- Blusztajn JK, Lopez Gonzalez-Coviella I, Logue M, Growdon JH, Wurtman RJ: Levels of phospholipid catabolic intermediates, glycerophosphocholine and glycerophosphoethanolamine, are elevated in brains of Alzheimer's disease but not of Down's syndrome patients. Brain Res. 1990 Dec 17;536(1-2):240-4. [PubMed:2150771 ]
- Mandal PK, McClure RJ, Pettegrew JW: Interactions of Abeta(1-40) with glycerophosphocholine and intact erythrocyte membranes: fluorescence and circular dichroism studies. Neurochem Res. 2004 Dec;29(12):2273-9. [PubMed:15672550 ]
- Pomfret EA, daCosta KA, Schurman LL, Zeisel SH: Measurement of choline and choline metabolite concentrations using high-pressure liquid chromatography and gas chromatography-mass spectrometry. Anal Biochem. 1989 Jul;180(1):85-90. [PubMed:2817347 ]
- Nitsch RM, Blusztajn JK, Pittas AG, Slack BE, Growdon JH, Wurtman RJ: Evidence for a membrane defect in Alzheimer disease brain. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1671-5. [PubMed:1311847 ]
- Miller BL, Read S, Tang C, Jenden D: Differences in red blood cell choline and lipid-bound choline between patients with Alzheimer disease and control subjects. Neurobiol Aging. 1991 Jan-Feb;12(1):61-4. [PubMed:2002884 ]
- Jensen JE, Drost DJ, Menon RS, Williamson PC: In vivo brain (31)P-MRS: measuring the phospholipid resonances at 4 Tesla from small voxels. NMR Biomed. 2002 Aug;15(5):338-47. [PubMed:12203225 ]
- Zhang Q, Southall MD, Mezsick SM, Johnson C, Murphy RC, Konger RL, Travers JB: Epidermal peroxisome proliferator-activated receptor gamma as a target for ultraviolet B radiation. J Biol Chem. 2005 Jan 7;280(1):73-9. Epub 2004 Oct 29. [PubMed:15516334 ]
- Fallbrook A, Turenne SD, Mamalias N, Kish SJ, Ross BM: Phosphatidylcholine and phosphatidylethanolamine metabolites may regulate brain phospholipid catabolism via inhibition of lysophospholipase activity. Brain Res. 1999 Jul 10;834(1-2):207-10. [PubMed:10407117 ]
- Blank ML, Smith ZL, Fitzgerald V, Snyder F: The CoA-independent transacylase in PAF biosynthesis: tissue distribution and molecular species selectivity. Biochim Biophys Acta. 1995 Feb 9;1254(3):295-301. [PubMed:7857969 ]
- Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. [PubMed:20044567 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
| Show more...
|---|