| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-05-30 20:55:55 UTC |
|---|
| HMDB ID | HMDB0000251 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Taurine |
|---|
| Description | Taurine is a sulfur amino acid like methionine, cystine, cysteine, and homocysteine. It is a lesser-known amino acid because it is not incorporated into the structural building blocks of protein. Yet taurine is an essential amino acid in pre-term and newborn infants of humans and many other species. Adults can synthesize their own taurine, yet are probably dependent, in part, on dietary taurine. Taurine is abundant in the brain, heart, breast, gallbladder, and kidney and has important roles in health and disease in these organs. Taurine has many diverse biological functions including serving as a neurotransmitter in the brain, a stabilizer of cell membranes, and a facilitator in the transport of ions such as sodium, potassium, calcium, and magnesium. Taurine is highly concentrated in animal and fish protein, which are good sources of dietary taurine. It can be synthesized by the body from cysteine when vitamin B6 is present. Deficiency of taurine occurs in premature infants, neonates fed formula milk, and various disease states. Several inborn errors of taurine metabolism have been described. Perry syndrome is an unusual neuropsychiatric disorder inherited in an autosomal dominant fashion through three generations of a family. Symptoms began late in the fifth decade in 6 affected persons and death occurred after 4 to 6 years. The earliest and most prominent symptom was mental depression that was not responsive to antidepressant drugs or electroconvulsive therapy. Sleep disturbances, exhaustion, and marked weight loss were features. Parkinsonism developed later, and respiratory failure occurred terminally (OMIM: 168605 ). Hypertaurinuric cardiomyopathy describes congestive cardiomyopathy and markedly elevated urinary taurine levels (about 5 times normal). Other family members had late or holosystolic mitral valve prolapse and elevated urinary taurine values (about 2.5 times normal). In two with mitral valve prolapse, congestive cardiomyopathy eventually developed while the amounts of urinary taurine doubled (OMIM: 145350 ). Taurine, after GABA, is the second most important inhibitory neurotransmitter in the brain. Its inhibitory effect is one source of taurine's anticonvulsant and antianxiety properties. It also lowers glutamic acid in the brain, and preliminary clinical trials suggest taurine may be useful in some forms of epilepsy. Taurine in the brain is usually associated with zinc or manganese. The amino acids alanine and glutamic acid, as well as pantothenic acid, inhibit taurine metabolism while vitamins A and B6, zinc, and manganese help build taurine. Cysteine and B6 are the nutrients most directly involved in taurine synthesis. Taurine levels have been found to decrease significantly in many depressed patients. One reason that the findings are not entirely clear is that taurine is often elevated in the blood of epileptics who need it. It is often difficult to distinguish compensatory changes in human biochemistry from true metabolic or deficiency disease. Low levels of taurine are found in retinitis pigmentosa. Taurine deficiency in experimental animals produces degeneration of light-sensitive cells. Therapeutic applications of taurine to eye disease are likely to be forthcoming. Taurine has many important metabolic roles. Supplements can stimulate prolactin and insulin release. The parathyroid gland makes a peptide hormone called glutataurine (glutamic acid-taurine), which further demonstrates taurine's role in endocrinology. Taurine increases bilirubin and cholesterol excretion in bile, critical to normal gallbladder function. It seems to inhibit the effect of morphine and potentiates the effects of opiate antagonists. Low plasma taurine levels have been found in a variety of conditions, i.e. depression, hypertension, hypothyroidism, gout, institutionalized patients, infertility, obesity, kidney failure, and others (http://www.dcnutrition.com/AminoAcids/). Moreover, taurine is found to be associated with maple syrup urine disease, which is an inborn error of metabolism. | Read more...
|---|
| Structure | InChI=1S/C2H7NO3S/c3-1-2-7(4,5)6/h1-3H2,(H,4,5,6) |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Aminoethanesulfonic acid | ChEBI | | 2-Aminoethyl sulfonate | ChEBI | | Aminoethylsulfonic acid | ChEBI | | beta-Aminoethylsulfonic acid | ChEBI | | 2-Aminoethanesulfonate | Generator | | 2-Aminoethanesulphonate | Generator | | 2-Aminoethanesulphonic acid | Generator | | 2-Aminoethyl sulfonic acid | Generator | | 2-Aminoethyl sulphonate | Generator | | 2-Aminoethyl sulphonic acid | Generator | | Aminoethylsulfonate | Generator | | Aminoethylsulphonate | Generator | | Aminoethylsulphonic acid | Generator | | b-Aminoethylsulfonate | Generator | | b-Aminoethylsulfonic acid | Generator | | b-Aminoethylsulphonate | Generator | | b-Aminoethylsulphonic acid | Generator | | beta-Aminoethylsulfonate | Generator | | beta-Aminoethylsulphonate | Generator | | beta-Aminoethylsulphonic acid | Generator | | Β-aminoethylsulfonate | Generator | | Β-aminoethylsulfonic acid | Generator | | Β-aminoethylsulphonate | Generator | | Β-aminoethylsulphonic acid | Generator | | 1-Aminoethane-2-sulfonate | HMDB | | 1-Aminoethane-2-sulfonic acid | HMDB | | 2-Aminoethylsulfonate | HMDB | | 2-Aminoethylsulfonic acid | HMDB | | 2-Sulfoethylamine | HMDB | | Taurine hydrochloride | HMDB | | Taurine zinc salt (2:1) | HMDB | | Taurine, monopotassium salt | HMDB | | Taufon | HMDB | | Tauphon | HMDB |
| Show more...
|---|
| Chemical Formula | C2H7NO3S |
|---|
| Average Molecular Weight | 125.147 |
|---|
| Monoisotopic Molecular Weight | 125.014663785 |
|---|
| IUPAC Name | 2-aminoethane-1-sulfonic acid |
|---|
| Traditional Name | taurine |
|---|
| CAS Registry Number | 107-35-7 |
|---|
| SMILES | NCCS(O)(=O)=O |
|---|
| InChI Identifier | InChI=1S/C2H7NO3S/c3-1-2-7(4,5)6/h1-3H2,(H,4,5,6) |
|---|
| InChI Key | XOAAWQZATWQOTB-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as organosulfonic acids. Organosulfonic acids are compounds containing the sulfonic acid group, which has the general structure RS(=O)2OH (R is not a hydrogen atom). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic sulfonic acids and derivatives |
|---|
| Sub Class | Organosulfonic acids and derivatives |
|---|
| Direct Parent | Organosulfonic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alkanesulfonic acid
- Sulfonyl
- Organosulfonic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organosulfur compound
- Organonitrogen compound
- Primary aliphatic amine
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 300 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 80.7 mg/mL | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | | Show more...
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Taurine,1TMS,isomer #1 | C[Si](C)(C)OS(=O)(=O)CCN | 1360.8 | Semi standard non polar | 33892256 | | Taurine,1TMS,isomer #1 | C[Si](C)(C)OS(=O)(=O)CCN | 1203.6 | Standard non polar | 33892256 | | Taurine,1TMS,isomer #1 | C[Si](C)(C)OS(=O)(=O)CCN | 2301.2 | Standard polar | 33892256 | | Taurine,1TMS,isomer #2 | C[Si](C)(C)NCCS(=O)(=O)O | 1497.7 | Semi standard non polar | 33892256 | | Taurine,1TMS,isomer #2 | C[Si](C)(C)NCCS(=O)(=O)O | 1243.5 | Standard non polar | 33892256 | | Taurine,1TMS,isomer #2 | C[Si](C)(C)NCCS(=O)(=O)O | 2483.3 | Standard polar | 33892256 | | Taurine,2TMS,isomer #1 | C[Si](C)(C)NCCS(=O)(=O)O[Si](C)(C)C | 1540.6 | Semi standard non polar | 33892256 | | Taurine,2TMS,isomer #1 | C[Si](C)(C)NCCS(=O)(=O)O[Si](C)(C)C | 1435.2 | Standard non polar | 33892256 | | Taurine,2TMS,isomer #1 | C[Si](C)(C)NCCS(=O)(=O)O[Si](C)(C)C | 1911.2 | Standard polar | 33892256 | | Taurine,2TMS,isomer #2 | C[Si](C)(C)N(CCS(=O)(=O)O)[Si](C)(C)C | 1615.4 | Semi standard non polar | 33892256 | | Taurine,2TMS,isomer #2 | C[Si](C)(C)N(CCS(=O)(=O)O)[Si](C)(C)C | 1569.6 | Standard non polar | 33892256 | | Taurine,2TMS,isomer #2 | C[Si](C)(C)N(CCS(=O)(=O)O)[Si](C)(C)C | 2257.4 | Standard polar | 33892256 | | Taurine,3TMS,isomer #1 | C[Si](C)(C)OS(=O)(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1691.7 | Semi standard non polar | 33892256 | | Taurine,3TMS,isomer #1 | C[Si](C)(C)OS(=O)(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1721.0 | Standard non polar | 33892256 | | Taurine,3TMS,isomer #1 | C[Si](C)(C)OS(=O)(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1856.3 | Standard polar | 33892256 | | Taurine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)(=O)CCN | 1610.0 | Semi standard non polar | 33892256 | | Taurine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)(=O)CCN | 1513.1 | Standard non polar | 33892256 | | Taurine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)(=O)CCN | 2465.1 | Standard polar | 33892256 | | Taurine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCS(=O)(=O)O | 1739.5 | Semi standard non polar | 33892256 | | Taurine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCS(=O)(=O)O | 1557.3 | Standard non polar | 33892256 | | Taurine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCS(=O)(=O)O | 2600.5 | Standard polar | 33892256 | | Taurine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C | 2007.6 | Semi standard non polar | 33892256 | | Taurine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C | 2003.1 | Standard non polar | 33892256 | | Taurine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C | 2082.9 | Standard polar | 33892256 | | Taurine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C | 2097.0 | Semi standard non polar | 33892256 | | Taurine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C | 2091.3 | Standard non polar | 33892256 | | Taurine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C | 2299.9 | Standard polar | 33892256 | | Taurine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2354.3 | Semi standard non polar | 33892256 | | Taurine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2517.2 | Standard non polar | 33892256 | | Taurine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2121.5 | Standard polar | 33892256 |
| Show more...
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-0032-1901000000-5373a9d32fa1f29b8012 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0032-0901000000-f7cf5a7ef7741fe71454 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00di-9802000000-c315203efd199b1871e7 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Non-derivatized) | splash10-0032-1901000000-5373a9d32fa1f29b8012 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Non-derivatized) | splash10-0032-0901000000-f7cf5a7ef7741fe71454 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Non-derivatized) | splash10-00di-9802000000-c315203efd199b1871e7 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Non-derivatized) | splash10-0f9t-0902000000-41e2bea3bce85bb00386 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Taurine GC-EI-TOF (Non-derivatized) | splash10-0f9t-0902000000-41e2bea3bce85bb00386 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurine GC-MS (Non-derivatized) - 70eV, Positive | splash10-000x-9000000000-fc8f496bf8d00c37dd84 | 2016-09-22 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-0036-9000000000-a96a1a8b53b1b556f6ca | 2014-09-20 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-006t-0890202100-ef2de9b47cafa707f789 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-004i-9300000000-530438ccab9f22503af5 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-00di-0900000000-ad642e440d924dfd8e20 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-006t-0097000000-24cfd722ea30b6a3b22d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-006t-0790202100-45d57f0101d4cefbc981 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-004i-9300000000-bd1f04d82f9e675f2303 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-00di-0900000000-d411fff703670832e445 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative-QTOF | splash10-006t-0098000000-c86b66ff71cff513520f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-00di-0900000000-31f44189a2671cf5d2e9 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-00e9-6900000000-cc7fe1badfe13959526d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-001i-9000000000-ae6465f92e91cfad7276 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-001i-9000000000-af0ea33a51fe79abee00 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-001i-9000000000-27d6c7ae55f73bb21b3c | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ , negative-QTOF | splash10-00di-0900000000-31f44189a2671cf5d2e9 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ , negative-QTOF | splash10-00e9-6900000000-cc7fe1badfe13959526d | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ , negative-QTOF | splash10-001i-9000000000-ae6465f92e91cfad7276 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ , negative-QTOF | splash10-001i-9000000000-af0ea33a51fe79abee00 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-QQ , negative-QTOF | splash10-001i-9000000000-27d6c7ae55f73bb21b3c | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT , negative-QTOF | splash10-004i-9300000000-530438ccab9f22503af5 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT , negative-QTOF | splash10-00di-0900000000-ad642e440d924dfd8e20 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT , negative-QTOF | splash10-006t-0097000000-e6d9cd15ddc56b10a1c6 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT , negative-QTOF | splash10-004i-9300000000-bd1f04d82f9e675f2303 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT , negative-QTOF | splash10-00di-0900000000-d411fff703670832e445 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine LC-ESI-ITFT , negative-QTOF | splash10-006t-0098000000-c86b66ff71cff513520f | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurine , negative-QTOF | splash10-00di-4900000000-9793e5415fbc98c19205 | 2017-09-14 | HMDB team, MONA | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Experimental 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | 2021-10-10 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
| Show more...
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Biospecimen Locations | - Bile
- Blood
- Breast Milk
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Urine
|
|---|
| Tissue Locations | - Brain
- Epidermis
- Erythrocyte
- Fibroblasts
- Intestine
- Kidney
- Leukocyte
- Liver
- Neuron
- Pancreas
- Placenta
- Platelet
- Prostate
- Retina
- Skeletal Muscle
|
|---|
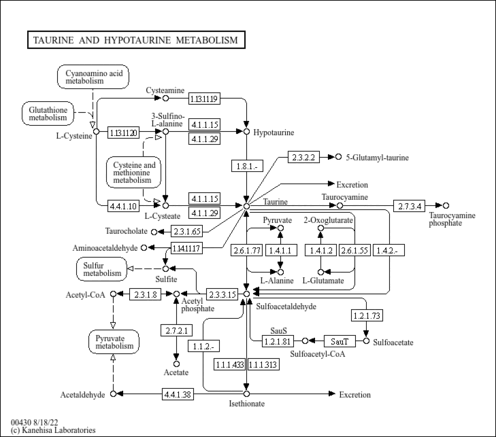
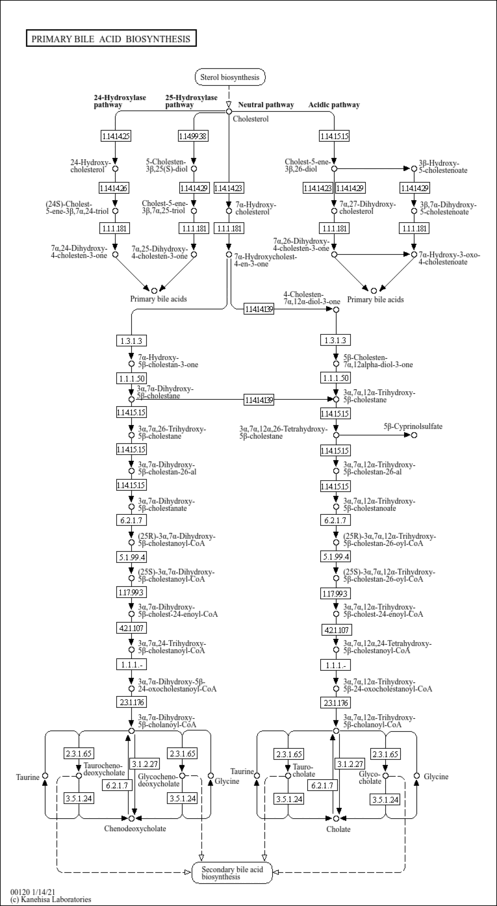
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Bile | Detected and Quantified | >10 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 0-360 uM | Infant (0-1 year old) | Both | Normal | | details | | Blood | Detected and Quantified | 55.0 (42.0-69.0) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 61 +/- 17 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 42 +/- 12 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 104.0 +/- 62.0 uM | Children (1-13 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 162.0 +/- 60.0 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 141.0 +/- 57.0 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 140 +/- 40 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 70.00-170.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 93.0 +/- 35.7 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 152.1(113.7-179.8) uM | Children (1-13 uears old) | Both | Normal | | details | | Blood | Detected and Quantified | 91.0 +/- 3.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 61.0 +/- 17.0 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 42.0 +/- 12.0 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 19-265 uM | Newborn (0-30 days old) | Both | Normal | | details | | Blood | Detected and Quantified | 19-190 uM | Infant (1 - 3 months old) | Both | Normal | | details | | Blood | Detected and Quantified | 10-97 uM | Children (3 months - 6 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 0-240 uM | Children (6 - 18 years old) | Both | Normal | | details | | Breast Milk | Detected and Quantified | 190 +/- 66.6 uM | Adult (>18 years old) | Female | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 6.6 +/- 1.7 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 8.24 (5.48-11.0) uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 13.4 +/- 2.7 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 8.4 +/- 2.0 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 5.5 +/- 1.5 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0-16 uM | Children (0 - 10 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0-14 uM | Adolescent (>11 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 5.87 +/- 0.63 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 5.3 +/- 2.2 uM | Not Specified | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected and Quantified | 140 +/- 290 nmol/g wet feces | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Normal | | details | | Feces | Detected and Quantified | 10 +/- 10 nmol/g wet feces | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 40.1 +/- 24.5 uM | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected and Quantified | 105.56 +/- 56.53 uM | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 57.8 +/- 24.3 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 52 (10-268) uM | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected and Quantified | 66 (1-171) uM | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected and Quantified | 45 (<1-184) uM | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected and Quantified | 25 (<1-69) uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 1-418 uM | Adult (>18 years old) | Male | normal | | details | | Saliva | Detected and Quantified | 1-56 uM | Adult (>18 years old) | Male | normal | | details | | Urine | Detected and Quantified | 350.49 +/- 192.20 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 463.55 +/- 214.81 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 520.08 +/- 293.96 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 54.7 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 74.64-2841 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 1114 +/- 761.3 umol/mmol creatinine | Newborn (0-30 days old) | Female | Normal | | details | | Urine | Detected and Quantified | 6.45-252.82 umol/mmol creatinine | Adolescent (13-18 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 1042 +/- 591.5 umol/mmol creatinine | Newborn (0-30 days old) | Male | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 4.00-159.98 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 2.71-173.18 umol/mmol creatinine | Adult (>18 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 4.18-938.73 umol/mmol creatinine | Infant (0-1 year old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 8.37 - 293.50 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 5.65 - 124.48 umol/mmol creatinine | Infant (1 - 6 months old) | Both | Normal | | details | | Urine | Detected and Quantified | 9.72 - 158.62 umol/mmol creatinine | Infant (6 months - <1 year old) | Both | Normal | | details | | Urine | Detected and Quantified | 13.57 - 205.77 umol/mmol creatinine | Children (1 - 2 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 14.81 - 259.47 umol/mmol creatinine | Children (2 - 4 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 20.01 - 298.59 umol/mmol creatinine | Children (4 - 13 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 18.65 - 233.13 umol/mmol creatinine | Adolescent (13 - 21 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 18.65 - 233.13 umol/mmol creatinine | Adult (>21 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 90.239 +/- 61.212 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 57.42 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 81 (13-251) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 7.24-368.20 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 1107.39 (537.18-1691.0) umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 1.6 +/- 1.6 umol/mmol creatinine | Infant (1 - 6 months old) | Both | Normal | | details | | Urine | Detected and Quantified | 2.5 +/-3.2 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 3.2 +/- 3.1 umol/mmol creatinine | Infant (6 months - <1 year old) | Both | Normal | | details | | Urine | Detected and Quantified | 36.07 +/- 29.16 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 60.45 (17.78-103.11) umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | - Geigy Scientific ...
- West Cadwell, N.J...
- Basel, Switzerlan...
| details | | Urine | Detected and Quantified | 8.60-398.062 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 63.2 (21.1-105.0) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 250-910 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 5.66-232.0048 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details |
| Show more...
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 188 uM | Infant (0-1 year old) | Male | Sulfite oxidase deficiency | | details | | Blood | Detected and Quantified | 117.0 +/- 7.0 uM | Adult (>18 years old) | Both | Heart failure | | details | | Blood | Detected and Quantified | 67 +/- 20 uM | Adult (>18 years old) | Male | Schizophrenia | | details | | Blood | Detected and Quantified | 68 +/- 14 uM | Adult (>18 years old) | Female | Schizophrenia | | details | | Blood | Detected and Quantified | 182.7(150.9-226.9) uM | Children (1-13 uears old) | Both | Environmental enteric dysfunction | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 5.80 (5.30-6.30) uM | Adult (>18 years old) | Both | Epilepsy | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 6.49 (4.60-8.38) uM | Adult (>18 years old) | Both | Parkinson's Disease | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 8.0 +/- 4.3 uM | Children (1-13 years old) | Not Specified | Leukemia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 10.4 +/- 11.1 uM | Children (1-13 years old) | Not Specified | Leukemia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 5.0 +/- 0.86 uM | Adult (>18 years old) | Both | Schizophrenia | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Gout | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal Cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | ankylosing spondylitis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | rheumatoid arthritis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Irritable bowel syndrome | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Both | Crohns disease | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Both | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Crohns disease | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Unclassified IBD | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Oral cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Breast cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Pancreatic cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Periodontal diseases | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Epithelial ovarian cancer | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Schizophrenia | | details | | Urine | Detected and Quantified | 58.393 +/- 30.929 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Eosinophilic esophagitis | | details | | Urine | Detected and Quantified | 0.0027 - 0.075 umol/mmol creatinine | Adult (>18 years old) | Both | ADPKD | | details | | Urine | Detected and Quantified | 67 +/- 50 umol/mmol creatinine | Adult (>18 years old) | Both | Lung cancer | | details | | Urine | Detected and Quantified | 466 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Molybdenum cofactor deficiency | | details | | Urine | Detected and Quantified | 1261 umol/mmol creatinine | Newborn (0-30 days old) | Not Specified | Molybdenum cofactor deficiency | | details | | Urine | Detected and Quantified | 280000000 umol/mmol creatinine | Children (1-13 years old) | Female | Sulfite oxidase deficiency | | details | | Urine | Detected and Quantified | 121.6 +/- 3.5 umol/mmol creatinine | Adult (>18 years old) | Both | 3-Hydroxy-3- methylglutaryl-CoA lyase (HL) deficency | | details | | Urine | Detected and Quantified | 72.9 +/- 10.7 umol/mmol creatinine | Adult (>18 years old) | Both | Propionic acidemia | | details | | Urine | Detected and Quantified | 98.3 +/- 7.1 umol/mmol creatinine | Adult (>18 years old) | Both | Maple syrup urine disease | | details | | Urine | Detected and Quantified | 49.4 +/- 1.4 umol/mmol creatinine | Adult (>18 years old) | Both | Aminoaciduria | | details |
| Show more...
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Heart failure |
|---|
- Norrelund H, Wiggers H, Halbirk M, Frystyk J, Flyvbjerg A, Botker HE, Schmitz O, Jorgensen JO, Christiansen JS, Moller N: Abnormalities of whole body protein turnover, muscle metabolism and levels of metabolic hormones in patients with chronic heart failure. J Intern Med. 2006 Jul;260(1):11-21. [PubMed:16789974 ]
| | Sulfite oxidase deficiency, ISOLATED |
|---|
- Rocha S, Ferreira AC, Dias AI, Vieira JP, Sequeira S: Sulfite oxidase deficiency--an unusual late and mild presentation. Brain Dev. 2014 Feb;36(2):176-9. doi: 10.1016/j.braindev.2013.01.013. Epub 2013 Feb 27. [PubMed:23452914 ]
- Choong T. et al. (2010). Clinical and Laboratory Barriers to the Timely Diagnosis of Sulphite Oxidase Deficiency. Proceedings of Singapore Healthcare, 19(2), 94-100.. Proceedings of Singapore Healthcare.
| | Epilepsy |
|---|
- Rainesalo S, Keranen T, Palmio J, Peltola J, Oja SS, Saransaari P: Plasma and cerebrospinal fluid amino acids in epileptic patients. Neurochem Res. 2004 Jan;29(1):319-24. [PubMed:14992292 ]
| | Parkinson's disease |
|---|
- Engelborghs S, Marescau B, De Deyn PP: Amino acids and biogenic amines in cerebrospinal fluid of patients with Parkinson's disease. Neurochem Res. 2003 Aug;28(8):1145-50. [PubMed:12834252 ]
| | Leukemia |
|---|
- Peng CT, Wu KH, Lan SJ, Tsai JJ, Tsai FJ, Tsai CH: Amino acid concentrations in cerebrospinal fluid in children with acute lymphoblastic leukemia undergoing chemotherapy. Eur J Cancer. 2005 May;41(8):1158-63. Epub 2005 Apr 14. [PubMed:15911239 ]
| | Schizophrenia |
|---|
- Do KQ, Lauer CJ, Schreiber W, Zollinger M, Gutteck-Amsler U, Cuenod M, Holsboer F: gamma-Glutamylglutamine and taurine concentrations are decreased in the cerebrospinal fluid of drug-naive patients with schizophrenic disorders. J Neurochem. 1995 Dec;65(6):2652-62. [PubMed:7595563 ]
- Bjerkenstedt L, Edman G, Hagenfeldt L, Sedvall G, Wiesel FA: Plasma amino acids in relation to cerebrospinal fluid monoamine metabolites in schizophrenic patients and healthy controls. Br J Psychiatry. 1985 Sep;147:276-82. [PubMed:2415198 ]
- Cai HL, Li HD, Yan XZ, Sun B, Zhang Q, Yan M, Zhang WY, Jiang P, Zhu RH, Liu YP, Fang PF, Xu P, Yuan HY, Zhang XH, Hu L, Yang W, Ye HS: Metabolomic analysis of biochemical changes in the plasma and urine of first-episode neuroleptic-naive schizophrenia patients after treatment with risperidone. J Proteome Res. 2012 Aug 3;11(8):4338-50. doi: 10.1021/pr300459d. Epub 2012 Jul 26. [PubMed:22800120 ]
| | Irritable bowel syndrome |
|---|
- Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A: Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011 Sep 2;10(9):4208-18. doi: 10.1021/pr2003598. Epub 2011 Aug 8. [PubMed:21761941 ]
| | Ulcerative colitis |
|---|
- Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A: Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011 Sep 2;10(9):4208-18. doi: 10.1021/pr2003598. Epub 2011 Aug 8. [PubMed:21761941 ]
- Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V: Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2017 Mar 1;11(3):321-334. doi: 10.1093/ecco-jcc/jjw158. [PubMed:27609529 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Crohn's disease |
|---|
- Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V: Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2017 Mar 1;11(3):321-334. doi: 10.1093/ecco-jcc/jjw158. [PubMed:27609529 ]
| | Gout |
|---|
- Shao T, Shao L, Li H, Xie Z, He Z, Wen C: Combined Signature of the Fecal Microbiome and Metabolome in Patients with Gout. Front Microbiol. 2017 Feb 21;8:268. doi: 10.3389/fmicb.2017.00268. eCollection 2017. [PubMed:28270806 ]
| | Rheumatoid arthritis |
|---|
- Tie-juan ShaoZhi-xing HeZhi-jun XieHai-chang LiMei-jiao WangCheng-ping Wen. Characterization of ankylosing spondylitis and rheumatoid arthritis using 1H NMR-based metabolomics of human fecal extracts. Metabolomics. April 2016, 12:70 [Link]
| | Perillyl alcohol administration for cancer treatment |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Pancreatic cancer |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Periodontal disease |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Lung Cancer |
|---|
- Stretch C, Eastman T, Mandal R, Eisner R, Wishart DS, Mourtzakis M, Prado CM, Damaraju S, Ball RO, Greiner R, Baracos VE: Prediction of skeletal muscle and fat mass in patients with advanced cancer using a metabolomic approach. J Nutr. 2012 Jan;142(1):14-21. doi: 10.3945/jn.111.147751. Epub 2011 Dec 7. [PubMed:22157537 ]
| | Autosomal dominant polycystic kidney disease |
|---|
- Gronwald W, Klein MS, Zeltner R, Schulze BD, Reinhold SW, Deutschmann M, Immervoll AK, Boger CA, Banas B, Eckardt KU, Oefner PJ: Detection of autosomal dominant polycystic kidney disease by NMR spectroscopic fingerprinting of urine. Kidney Int. 2011 Jun;79(11):1244-53. doi: 10.1038/ki.2011.30. Epub 2011 Mar 9. [PubMed:21389975 ]
| | Propionic acidemia |
|---|
- Gronwald W, Klein MS, Kaspar H, Fagerer SR, Nurnberger N, Dettmer K, Bertsch T, Oefner PJ: Urinary metabolite quantification employing 2D NMR spectroscopy. Anal Chem. 2008 Dec 1;80(23):9288-97. doi: 10.1021/ac801627c. [PubMed:19551947 ]
| | Maple syrup urine disease |
|---|
- Gronwald W, Klein MS, Kaspar H, Fagerer SR, Nurnberger N, Dettmer K, Bertsch T, Oefner PJ: Urinary metabolite quantification employing 2D NMR spectroscopy. Anal Chem. 2008 Dec 1;80(23):9288-97. doi: 10.1021/ac801627c. [PubMed:19551947 ]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
| | Molybdenum cofactor deficiency |
|---|
- van Gennip AH, Abeling NG, Stroomer AE, Overmars H, Bakker HD: The detection of molybdenum cofactor deficiency: clinical symptomatology and urinary metabolite profile. J Inherit Metab Dis. 1994;17(1):142-5. [PubMed:8051926 ]
|
|
|---|
| Associated OMIM IDs | - 272300 (Sulfite oxidase deficiency, ISOLATED)
- 168600 (Parkinson's disease)
- 181500 (Schizophrenia)
- 114500 (Colorectal cancer)
- 266600 (Crohn's disease)
- 138900 (Gout)
- 180300 (Rheumatoid arthritis)
- 260350 (Pancreatic cancer)
- 170650 (Periodontal disease)
- 211980 (Lung Cancer)
- 601313 (Autosomal dominant polycystic kidney disease)
- 606054 (Propionic acidemia)
- 248600 (Maple syrup urine disease)
- 610247 (Eosinophilic esophagitis)
|
|---|
| External Links |
|---|
| DrugBank ID | DB01956 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB003191 |
|---|
| KNApSAcK ID | C00048188 |
|---|
| Chemspider ID | 1091 |
|---|
| KEGG Compound ID | C00245 |
|---|
| BioCyc ID | TAURINE |
|---|
| BiGG ID | 34373 |
|---|
| Wikipedia Link | Taurine |
|---|
| METLIN ID | 31 |
|---|
| PubChem Compound | 1123 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15891 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | TAUR |
|---|
| MarkerDB ID | MDB00000122 |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Hu, Libo; Zhu, Hui; Du, Da-Ming; Xu, Jiaxi. Efficient synthesis of taurine and structurally diverse substituted taurines from aziridines. Journal of Organic Chemistry (2007), 72(12), 4543-4546. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| General References | - Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Engelborghs S, Marescau B, De Deyn PP: Amino acids and biogenic amines in cerebrospinal fluid of patients with Parkinson's disease. Neurochem Res. 2003 Aug;28(8):1145-50. [PubMed:12834252 ]
- Hagenfeldt L, Bjerkenstedt L, Edman G, Sedvall G, Wiesel FA: Amino acids in plasma and CSF and monoamine metabolites in CSF: interrelationship in healthy subjects. J Neurochem. 1984 Mar;42(3):833-7. [PubMed:6198473 ]
- Peng CT, Wu KH, Lan SJ, Tsai JJ, Tsai FJ, Tsai CH: Amino acid concentrations in cerebrospinal fluid in children with acute lymphoblastic leukemia undergoing chemotherapy. Eur J Cancer. 2005 May;41(8):1158-63. Epub 2005 Apr 14. [PubMed:15911239 ]
- Cynober LA: Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition. 2002 Sep;18(9):761-6. [PubMed:12297216 ]
- Rainesalo S, Keranen T, Palmio J, Peltola J, Oja SS, Saransaari P: Plasma and cerebrospinal fluid amino acids in epileptic patients. Neurochem Res. 2004 Jan;29(1):319-24. [PubMed:14992292 ]
- Khan SA, Cox IJ, Hamilton G, Thomas HC, Taylor-Robinson SD: In vivo and in vitro nuclear magnetic resonance spectroscopy as a tool for investigating hepatobiliary disease: a review of H and P MRS applications. Liver Int. 2005 Apr;25(2):273-81. [PubMed:15780050 ]
- Vinton NE, Laidlaw SA, Ament ME, Kopple JD: Taurine concentrations in plasma, blood cells, and urine of children undergoing long-term total parenteral nutrition. Pediatr Res. 1987 Apr;21(4):399-403. [PubMed:3106924 ]
- Gonzalez-Quevedo A, Obregon F, Fernandez R, Santiesteban R, Serrano C, Lima L: Amino acid levels and ratios in serum and cerebrospinal fluid of patients with optic neuropathy in Cuba. Nutr Neurosci. 2001;4(1):51-62. [PubMed:11842876 ]
- Schneider SM, Joly F, Gehrardt MF, Badran AM, Myara A, Thuillier F, Coudray-Lucas C, Cynober L, Trivin F, Messing B: Taurine status and response to intravenous taurine supplementation in adults with short-bowel syndrome undergoing long-term parenteral nutrition: a pilot study. Br J Nutr. 2006 Aug;96(2):365-70. [PubMed:16923232 ]
- McCarty MF: Complementary vascular-protective actions of magnesium and taurine: a rationale for magnesium taurate. Med Hypotheses. 1996 Feb;46(2):89-100. [PubMed:8692051 ]
- Kopple JD, Vinton NE, Laidlaw SA, Ament ME: Effect of intravenous taurine supplementation on plasma, blood cell, and urine taurine concentrations in adults undergoing long-term parenteral nutrition. Am J Clin Nutr. 1990 Nov;52(5):846-53. [PubMed:2122710 ]
- McMahon GP, O'Kennedy R, Kelly MT: High-performance liquid chromatographic determination of taurine in human plasma using pre-column extraction and derivatization. J Pharm Biomed Anal. 1996 Jun;14(8-10):1287-94. [PubMed:8818047 ]
- Stover JF, Morganti-Kosmann MC, Lenzlinger PM, Stocker R, Kempski OS, Kossmann T: Glutamate and taurine are increased in ventricular cerebrospinal fluid of severely brain-injured patients. J Neurotrauma. 1999 Feb;16(2):135-42. [PubMed:10098958 ]
- Learn DB, Fried VA, Thomas EL: Taurine and hypotaurine content of human leukocytes. J Leukoc Biol. 1990 Aug;48(2):174-82. [PubMed:2370482 ]
- Miglis M, Wilder D, Reid T, Bakaltcheva I: Effect of taurine on platelets and the plasma coagulation system. Platelets. 2002 Feb;13(1):5-10. [PubMed:11918831 ]
- Axelson M, Ellis E, Mork B, Garmark K, Abrahamsson A, Bjorkhem I, Ericzon BG, Einarsson C: Bile acid synthesis in cultured human hepatocytes: support for an alternative biosynthetic pathway to cholic acid. Hepatology. 2000 Jun;31(6):1305-12. [PubMed:10827156 ]
- Hu S, Zhao X, Yin S, Meng J: [A study on the mechanism of taurine postponing the aging process of human fetal brain neural cells]. Wei Sheng Yan Jiu. 1997 Mar;26(2):98-101. [PubMed:10325611 ]
- Goodman HO, Shihabi Z, Oles KS: Antiepileptic drugs and plasma and platelet taurine in epilepsy. Epilepsia. 1989 Mar-Apr;30(2):201-7. [PubMed:2494044 ]
- Sturman JA, Messing JM, Rossi SS, Hofmann AF, Neuringer MD: Tissue taurine content and conjugated bile acid composition of rhesus monkey infants fed a human infant soy-protein formula with or without taurine supplementation for 3 months. Neurochem Res. 1988 Apr;13(4):311-6. [PubMed:3393260 ]
- Gonzalez-Quevedo A, Obregon F, Santiesteban Freixas R, Fernandez R, Lima L: [Amino acids as biochemical markers in epidemic and endemic optic neuropathies]. Rev Cubana Med Trop. 1998;50 Suppl:241-4. [PubMed:10349454 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
| Show more...
|---|