| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2006-08-13 07:51:07 UTC |
|---|
| Update Date | 2022-03-07 02:49:20 UTC |
|---|
| HMDB ID | HMDB0003944 |
|---|
| Secondary Accession Numbers | - HMDB0012970
- HMDB03944
- HMDB12970
|
|---|
| Metabolite Identification |
|---|
| Common Name | trans-2-Hexenoyl-CoA |
|---|
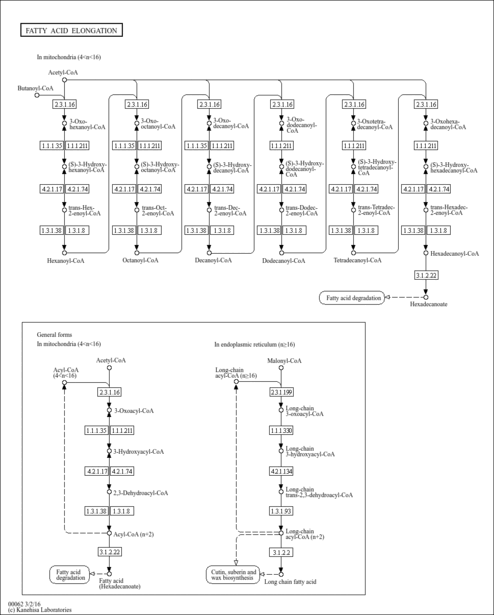
| Description | trans-Hexenoyl-CoA is an intermediate in fatty acid metabolism. Beta-oxidation occurs in both mitochondria and peroxisomes. Mitochondria catalyze the beta-oxidation of the bulk of short-, medium-, and long-chain fatty acids derived from diet, and this pathway constitutes the major process by which fatty acids are oxidized to generate energy. Peroxisomes are involved in the beta-oxidation chain shortening of long-chain and very-long-chain fatty acyl-coenzyme (CoAs), long-chain dicarboxylyl-CoAs, the CoA esters of eicosanoids, 2-methyl-branched fatty acyl-CoAs, and the CoA esters of the bile acid intermediates di- and trihydroxycoprostanoic acids, and in the process they generate H2O2. Long-chain and very-long-chain fatty acids (VLCFAs) are also metabolized by the cytochrome P450 CYP4A omega-oxidation system to dicarboxylic acids that serve as substrates for peroxisomal beta-oxidation. The peroxisomal beta-oxidation system consists of (a) a classical peroxisome proliferator-inducible pathway capable of catalyzing straight-chain acyl-CoAs by fatty acyl-CoA oxidase, L-bifunctional protein, and thiolase, and (b) a second noninducible pathway catalyzing the oxidation of 2-methyl-branched fatty acyl-CoAs by branched-chain acyl-CoA oxidase (pristanoyl-CoA oxidase/trihydroxycoprostanoyl-CoA oxidase), D-bifunctional protein, and sterol carrier protein (SCP)x. trans-Hexenoyl-CoA is the substrate of the enzymes enoyl-coenzyme A reductase, acyl-CoA oxidase [EC 1.3.99.2-1.3.3.6], acyl-CoA dehydrogenase, long-chain-acyl-CoA dehydrogenase [EC 1.3.99.3-1.3.99.13], and Oxidoreductases [EC 1.3.99.-]; trans-Hexenoyl-CoA is an intermediate in fatty acid elongation in mitochondria, being the substrate of the enzymes enoyl-CoA hydratase and long-chain-enoyl-CoA hydratase [EC 4.2.1.17-4.2.1.74]. (PMID: 11375435 ). |
|---|
| Structure | CCC\C=C\C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C27H44N7O17P3S/c1-4-5-6-7-18(36)55-11-10-29-17(35)8-9-30-25(39)22(38)27(2,3)13-48-54(45,46)51-53(43,44)47-12-16-21(50-52(40,41)42)20(37)26(49-16)34-15-33-19-23(28)31-14-32-24(19)34/h6-7,14-16,20-22,26,37-38H,4-5,8-13H2,1-3H3,(H,29,35)(H,30,39)(H,43,44)(H,45,46)(H2,28,31,32)(H2,40,41,42)/b7-6+/t16-,20-,21-,22?,26-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| (2E)-Hexenoyl-CoA | HMDB | | (2E)-Hexenoyl-coenzyme A | HMDB | | trans-2,3-Dehydrohexanoyl-CoA | HMDB | | trans-2,3-Dehydrohexanoyl-coenzyme A | HMDB | | trans-Hex-2-enoyl-CoA | HMDB | | trans-Hex-2-enoyl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C27H44N7O17P3S |
|---|
| Average Molecular Weight | 863.661 |
|---|
| Monoisotopic Molecular Weight | 863.172723243 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-{[({[(3-{[2-({2-[(2E)-hex-2-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}-3-hydroxy-2,2-dimethylpropoxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl)oxy]methyl}-4-hydroxyoxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-2-({[(3-{[2-({2-[(2E)-hex-2-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}-3-hydroxy-2,2-dimethylpropoxy(hydroxy)phosphoryl)oxy(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | 10018-93-6 |
|---|
| SMILES | CCC\C=C\C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C27H44N7O17P3S/c1-4-5-6-7-18(36)55-11-10-29-17(35)8-9-30-25(39)22(38)27(2,3)13-48-54(45,46)51-53(43,44)47-12-16-21(50-52(40,41)42)20(37)26(49-16)34-15-33-19-23(28)31-14-32-24(19)34/h6-7,14-16,20-22,26,37-38H,4-5,8-13H2,1-3H3,(H,29,35)(H,30,39)(H,43,44)(H,45,46)(H2,28,31,32)(H2,40,41,42)/b7-6+/t16-,20-,21-,22?,26-/m1/s1 |
|---|
| InChI Key | OINXHIBNZUUIMR-DOSZRKKKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as medium-chain 2-enoyl coas. These are organic compounds containing a coenzyme A substructure linked to a medium-chain 2-enoyl chain of 5 to 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Medium-chain 2-enoyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -2.181 | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 10V, Positive-QTOF | splash10-000i-1902000120-12f3a35f3f9ab291ff00 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 20V, Positive-QTOF | splash10-000i-1912000000-1f13bbf987a235864a7c | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 40V, Positive-QTOF | splash10-000i-2911000000-9a6ce611452163aa9b91 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 10V, Negative-QTOF | splash10-01sm-8930140550-852eb909c02343edb382 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 20V, Negative-QTOF | splash10-003r-6910010010-97a6c041da0e77ec8644 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 40V, Negative-QTOF | splash10-057i-6900100000-06a3c53b188324acdb55 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 10V, Positive-QTOF | splash10-03di-0100000090-0bad0b4bba13724e3e3c | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 20V, Positive-QTOF | splash10-000i-1901001380-548b3eab92846ec39a6b | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 40V, Positive-QTOF | splash10-0a4i-1119000000-014d8f50596dc71ea37c | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 10V, Negative-QTOF | splash10-03di-0000000090-05f215703b97646d7243 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 20V, Negative-QTOF | splash10-03gr-8400105950-dfb121a3f8352299d2ba | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - trans-2-Hexenoyl-CoA 40V, Negative-QTOF | splash10-0170-6103502920-9a1f4d2a3459bc629e5c | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum |
|
|---|