| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2006-08-13 07:50:32 UTC |
|---|
| Update Date | 2022-03-07 02:49:20 UTC |
|---|
| HMDB ID | HMDB0003943 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 3-Oxohexanoyl-CoA |
|---|
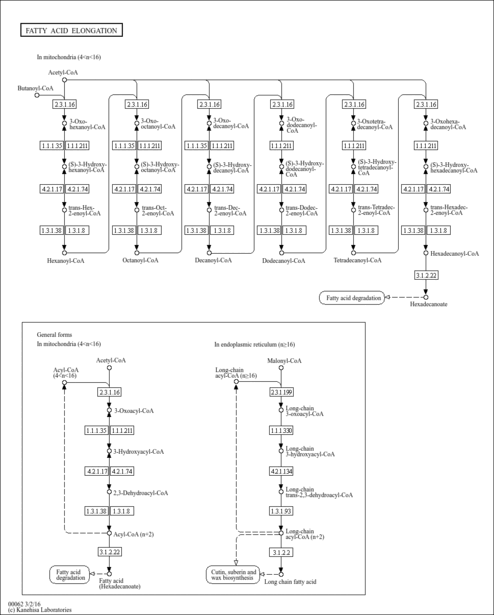
| Description | 3-Oxohexanoyl-CoA is an intermediate in Fatty acid elongation in mitochondria. 3-Oxohexanoyl-CoA is the 3rd to last step in the synthesis of Hexanoyl-CoA and is converted from Butanoyl-CoA via the enzyme acetyl-CoA acyltransferase 2 (EC 2.3.1.16). It is then converted to (S)-Hydroxyhexanoyl-CoA via the 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35). |
|---|
| Structure | CCCC(=O)CC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 InChI=1S/C27H44N7O18P3S/c1-4-5-15(35)10-18(37)56-9-8-29-17(36)6-7-30-25(40)22(39)27(2,3)12-49-55(46,47)52-54(44,45)48-11-16-21(51-53(41,42)43)20(38)26(50-16)34-14-33-19-23(28)31-13-32-24(19)34/h13-14,16,20-22,26,38-39H,4-12H2,1-3H3,(H,29,36)(H,30,40)(H,44,45)(H,46,47)(H2,28,31,32)(H2,41,42,43)/t16-,20-,21-,22?,26-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Ketohexanoyl coa. | HMDB | | 3-Ketohexanoyl coenzyme A. | HMDB | | 3-Oxohexanoyl-coenzyme A | HMDB | | Ketohexanoyl-CoA | HMDB | | Ketohexanoyl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C27H44N7O18P3S |
|---|
| Average Molecular Weight | 879.661 |
|---|
| Monoisotopic Molecular Weight | 879.167637865 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({[hydroxy(3-hydroxy-2,2-dimethyl-3-{[2-({2-[(3-oxohexanoyl)sulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}propoxy)phosphoryl]oxy})phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-[({hydroxy[hydroxy(3-hydroxy-2,2-dimethyl-3-{[2-({2-[(3-oxohexanoyl)sulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}propoxy)phosphoryl]oxyphosphoryl}oxy)methyl]oxolan-3-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | 19774-86-8 |
|---|
| SMILES | CCCC(=O)CC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 |
|---|
| InChI Identifier | InChI=1S/C27H44N7O18P3S/c1-4-5-15(35)10-18(37)56-9-8-29-17(36)6-7-30-25(40)22(39)27(2,3)12-49-55(46,47)52-54(44,45)48-11-16-21(51-53(41,42)43)20(38)26(50-16)34-14-33-19-23(28)31-13-32-24(19)34/h13-14,16,20-22,26,38-39H,4-12H2,1-3H3,(H,29,36)(H,30,40)(H,44,45)(H,46,47)(H2,28,31,32)(H2,41,42,43)/t16-,20-,21-,22?,26-/m1/s1 |
|---|
| InChI Key | NFOYYXQAVVYWKV-ZOGSZLKASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 3-oxo-acyl coas. These are organic compounds containing a 3-oxo acylated coenzyme A derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | 3-oxo-acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- 1,3-dicarbonyl compound
- Imidolactam
- Monosaccharide
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Ketone
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Amine
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -3.064 | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | | Show more...
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized | Show more...
|---|
| Spectra |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 10V, Positive-QTOF | splash10-000i-1901000120-68f6500665ed8e511dc7 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 20V, Positive-QTOF | splash10-000i-0912000000-54a826f27a4c6ba4efb3 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 40V, Positive-QTOF | splash10-000i-2911000000-0fb092f4177cad6035ba | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 10V, Negative-QTOF | splash10-01u0-4911030340-b74454836fd5a8474037 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 20V, Negative-QTOF | splash10-001i-3910110010-eb4a9cd58caa577306dc | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 40V, Negative-QTOF | splash10-057i-5900100000-70d1d4cf63d535a9c95c | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 10V, Negative-QTOF | splash10-004i-0000000190-507fcc0bdd5aad9b0deb | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 20V, Negative-QTOF | splash10-0170-5400103950-4e91268e6c531bd9d1f6 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 40V, Negative-QTOF | splash10-0f97-8102301920-4e5f2d7fbe293afed3a6 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 10V, Positive-QTOF | splash10-01q9-0000000190-b6306b6b1a0792384415 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 20V, Positive-QTOF | splash10-01ox-0100000190-c5adc0df23de84c08be6 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Oxohexanoyl-CoA 40V, Positive-QTOF | splash10-00di-0029000000-a01d7d00a2c64e79bbc0 | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum |
| Show more...
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Biospecimen Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | None |
|---|
| Associated OMIM IDs | None |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB023262 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 389508 |
|---|
| KEGG Compound ID | C05269 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 440610 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 27648 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | 3OHCOA |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| General References | Not Available |
|---|