| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:09 UTC |
|---|
| HMDB ID | HMDB0001358 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Retinal |
|---|
| Description | Retinal is a carotenoid constituent of visual pigments. It is the oxidized form of retinol which functions as the active component of the visual cycle. It is bound to the protein opsin forming the complex rhodopsin. When stimulated by visible light, the retinal component of the rhodopsin complex undergoes isomerization at the 11-position of the double bond to the cis-form; this is reversed in "dark" reactions to return to the native trans-configuration. |
|---|
| Structure | C\C(\C=C\C=C(/C)\C=C\C1=C(C)CCCC1(C)C)=C/C=O InChI=1S/C20H28O/c1-16(8-6-9-17(2)13-15-21)11-12-19-18(3)10-7-14-20(19,4)5/h6,8-9,11-13,15H,7,10,14H2,1-5H3/b9-6+,12-11+,16-8+,17-13+ |
|---|
| Synonyms | | Value | Source |
|---|
| all-trans-Retinaldehyde | ChEBI | | all-trans-Retinene | ChEBI | | all-trans-Vitamin a aldehyde | ChEBI | | Retinaldehyde | ChEBI | | Retinene | ChEBI | | Vitamin a aldehyde | ChEBI | | all-trans-Retinal | Kegg | | Aldehyde, vitamin a | HMDB | | Axerophthal | HMDB | | 11 cis Retinal | HMDB | | 11-cis-Retinal | HMDB | | 3,7-Dimethyl-9-(2,6,6-trimethyl-1-cyclohexen-1-yl)-2,4,6,8-nonatetraenal | HMDB | | all-e-Retinal | HMDB | | all-epsilon-Retinal | HMDB | | alpha-Retinene | HMDB | | e-Retinal | HMDB | | epsilon-Retinal | HMDB | | Retinene 1 | HMDB | | trans-Retinal | HMDB | | trans-Vitamin a aldehyde | HMDB | | Vitamin a1 aldehyde | HMDB |
|
|---|
| Chemical Formula | C20H28O |
|---|
| Average Molecular Weight | 284.4357 |
|---|
| Monoisotopic Molecular Weight | 284.214015518 |
|---|
| IUPAC Name | (2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-en-1-yl)nona-2,4,6,8-tetraenal |
|---|
| Traditional Name | retinaldehyde |
|---|
| CAS Registry Number | 116-31-4 |
|---|
| SMILES | C\C(\C=C\C=C(/C)\C=C\C1=C(C)CCCC1(C)C)=C/C=O |
|---|
| InChI Identifier | InChI=1S/C20H28O/c1-16(8-6-9-17(2)13-15-21)11-12-19-18(3)10-7-14-20(19,4)5/h6,8-9,11-13,15H,7,10,14H2,1-5H3/b9-6+,12-11+,16-8+,17-13+ |
|---|
| InChI Key | NCYCYZXNIZJOKI-OVSJKPMPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as retinoids. These are oxygenated derivatives of 3,7-dimethyl-1-(2,6,6-trimethylcyclohex-1-enyl)nona-1,3,5,7-tetraene and derivatives thereof. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Retinoids |
|---|
| Direct Parent | Retinoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Retinoid skeleton
- Diterpenoid
- Enal
- Alpha,beta-unsaturated aldehyde
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aldehyde
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 63 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized |
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Retinal GC-MS (Non-derivatized) - 70eV, Positive | splash10-014i-2290000000-df9d2aa7545cc60a54f9 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Retinal GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Retinal 40V, Positive-QTOF | splash10-014i-9700000000-c2e22000dfae0765c367 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Retinal 10V, Positive-QTOF | splash10-03g0-0950000000-bccf87de037f98873f41 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Retinal 20V, Positive-QTOF | splash10-01t9-1910000000-5fe7643c395769c68286 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Retinal 40V, Positive-QTOF | splash10-014i-9700000000-fca532f917595376d653 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 10V, Positive-QTOF | splash10-000i-0490000000-f543f08fbaf94a2e3f8e | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 20V, Positive-QTOF | splash10-000i-3940000000-74f087cc812b3715e7ac | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 40V, Positive-QTOF | splash10-0fri-9830000000-72a5d01c6181340ad986 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 10V, Negative-QTOF | splash10-001i-0090000000-bad35938b77f181c29b9 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 20V, Negative-QTOF | splash10-001i-0090000000-1aadad0770df07f98436 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 40V, Negative-QTOF | splash10-00ko-4690000000-f624d342f88456375b9c | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 10V, Positive-QTOF | splash10-00n3-1960000000-43f8c66f164764c37361 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 20V, Positive-QTOF | splash10-0609-3920000000-dbeab0a82b306761cf6a | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 40V, Positive-QTOF | splash10-052f-7900000000-d2bb7c1aeeb87d8833d2 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 10V, Negative-QTOF | splash10-0a59-0090000000-2ec2f8f314943441fa15 | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 20V, Negative-QTOF | splash10-0pb9-0190000000-312cebb17ccd0189ab4d | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Retinal 40V, Negative-QTOF | splash10-000i-1590000000-b2551eac481c0c930a4d | 2021-09-25 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 600 MHz, CDCl3, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane (predicted from logP)
|
|---|
| Biospecimen Locations | |
|---|
| Tissue Locations | - Adrenal Cortex
- Epidermis
- Fibroblasts
- Intestine
- Kidney
- Lung
- Neuron
- Pancreas
- Placenta
- Platelet
- Skeletal Muscle
- Spleen
- Testis
|
|---|
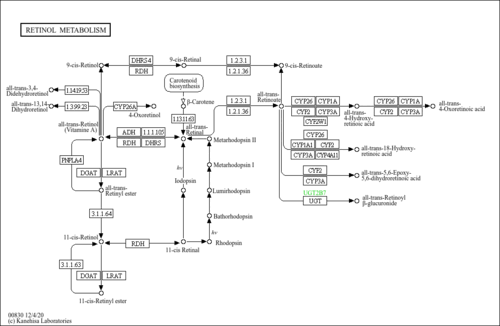
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 0.155 +/- 0.232 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | None |
|---|
| Associated OMIM IDs | None |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB022576 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00376 |
|---|
| BioCyc ID | RETINAL |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Retinal |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 638015 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17898 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | RETINAL |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Duhamel, Lucette; Duhamel, Pierre; Lecouve, Jean Pierre. A new synthesis of retinals. Journal of Chemical Research, Synopses (1986), (1), 34-5. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Chang M, Herbert WN: Retinal arteriolar occlusions following amniotic fluid embolism. Ophthalmology. 1984 Dec;91(12):1634-7. [PubMed:6521996 ]
- Nguyen QD, Van Do D, Feke GT, Demirjian ZN, Lashkari K: Heparin-induced antiheparin-platelet antibody associated with retinal venous thrombosis. Ophthalmology. 2003 Mar;110(3):600-3. [PubMed:12623829 ]
- Aarli JA, Mork SJ, Myrseth E, Larsen JL: Glioblastoma associated with multiple sclerosis: coincidence or induction? Eur Neurol. 1989;29(6):312-6. [PubMed:2691257 ]
- Wu WC, Lai CC, Liu JH, Singh T, Li LM, Peumans WJ, Van Damme EJ, Wu AM: Differential Binding to Glycotopes Among the Layers of Three Mammalian Retinal Neurons by Man-Containing N-linked Glycan, T(alpha) (Galbeta1-3GalNAcalpha1-), Tn (GalNAcalpha1-Ser/Thr) and I (beta)/II (beta) (Galbeta1-3/4GlcNAcbeta-) Reactive Lectins. Neurochem Res. 2006 May 23;. [PubMed:16718528 ]
- Jankowska R, Witkowska D, Porebska I, Kuropatwa M, Kurowska E, Gorczyca WA: Serum antibodies to retinal antigens in lung cancer and sarcoidosis. Pathobiology. 2004;71(6):323-8. [PubMed:15627843 ]
- Weiland JD, Liu W, Humayun MS: Retinal prosthesis. Annu Rev Biomed Eng. 2005;7:361-401. [PubMed:16004575 ]
- Fukushima A, Ueno H, Fujimoto S: Antigenic cross-reactivity between human T lymphotropic virus type I (HTLV-I) and retinal antigens recognized by T cells. Clin Exp Immunol. 1994 Mar;95(3):459-64. [PubMed:8137541 ]
- Bradbury MW, Lightman SL: The blood-brain interface. Eye (Lond). 1990;4 ( Pt 2):249-54. [PubMed:2199234 ]
- Bringmann A, Reichenbach A, Wiedemann P: Pathomechanisms of cystoid macular edema. Ophthalmic Res. 2004 Sep-Oct;36(5):241-9. [PubMed:15583429 ]
- Wu WC, Lai CC, Liu JH, Singh T, Li LM, Peumans WJ, Van Damme EJ, Wu AM: Differential binding to glycotopes among the layers of three mammalian retinal neurons by man-containing N-linked glycan, T(alpha) (Galbeta1-3GalNAcalpha1-), Tn (GalNAcalpha1-Ser/Thr) and I (beta)/II (beta) (Galbeta1-3/4GlcNAcbeta-) reactive lectins. Neurochem Res. 2006 May;31(5):619-28. Epub 2006 May 23. [PubMed:16770733 ]
- Yamashiro K, Tsujikawa A, Ishida S, Usui T, Kaji Y, Honda Y, Ogura Y, Adamis AP: Platelets accumulate in the diabetic retinal vasculature following endothelial death and suppress blood-retinal barrier breakdown. Am J Pathol. 2003 Jul;163(1):253-9. [PubMed:12819029 ]
- Ramirez JM, Ramirez AI, Salazar JJ, de Hoz R, Trivino A: Changes of astrocytes in retinal ageing and age-related macular degeneration. Exp Eye Res. 2001 Nov;73(5):601-15. [PubMed:11747361 ]
|
|---|